DeepRank-GNN: a graph neural network framework to learn patterns in protein-protein interfaces
- PMID: 36420989
- PMCID: PMC9805592
- DOI: 10.1093/bioinformatics/btac759
DeepRank-GNN: a graph neural network framework to learn patterns in protein-protein interfaces
Abstract
Motivation: Gaining structural insights into the protein-protein interactome is essential to understand biological phenomena and extract knowledge for rational drug design or protein engineering. We have previously developed DeepRank, a deep-learning framework to facilitate pattern learning from protein-protein interfaces using convolutional neural network (CNN) approaches. However, CNN is not rotation invariant and data augmentation is required to desensitize the network to the input data orientation which dramatically impairs the computation performance. Representing protein-protein complexes as atomic- or residue-scale rotation invariant graphs instead enables using graph neural networks (GNN) approaches, bypassing those limitations.
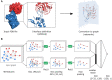
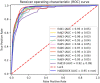
Results: We have developed DeepRank-GNN, a framework that converts protein-protein interfaces from PDB 3D coordinates files into graphs that are further provided to a pre-defined or user-defined GNN architecture to learn problem-specific interaction patterns. DeepRank-GNN is designed to be highly modularizable, easily customized and is wrapped into a user-friendly python3 package. Here, we showcase DeepRank-GNN's performance on two applications using a dedicated graph interaction neural network: (i) the scoring of docking poses and (ii) the discriminating of biological and crystal interfaces. In addition to the highly competitive performance obtained in those tasks as compared to state-of-the-art methods, we show a significant improvement in speed and storage requirement using DeepRank-GNN as compared to DeepRank.
Availability and implementation: DeepRank-GNN is freely available from https://github.com/DeepRank/DeepRank-GNN.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures
References
-
- Fout A. et al. (2017) Protein interface prediction using graph convolutional networks. In: Advances in Neural Information Processing Systems, Long Beach Conference Center, Los Angeles, USA.

