Addition of ROCK Inhibitors Alleviates Prostaglandin-Induced Inhibition of Adipogenesis in 3T3L-1 Spheroids
- PMID: 36421103
- PMCID: PMC9687287
- DOI: 10.3390/bioengineering9110702
Addition of ROCK Inhibitors Alleviates Prostaglandin-Induced Inhibition of Adipogenesis in 3T3L-1 Spheroids
Abstract
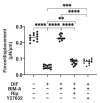
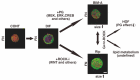
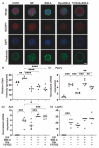
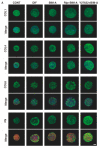
To elucidate the additive effects of the ROCK inhibitors (ROCK-i), ripasudil (Rip) and Y27632 on bimatoprost acid (BIM-A), a prostaglandin analog (PG), on adipose tissue, two- and three-dimensional (2D or 3D) cultures of 3T3-L1 cells, the most well characterized cells in the field of lipid research, were used. The cells were subjected to a variety of analyses including lipid staining, real-time cellular metabolic analysis, the mRNA expressions of genes related to adipogenesis and extracellular matrices (ECMs) as well as the sizes and physical properties of the 3D spheroids by a micro-squeezer. BIM-A induced strong inhibitory effects on most of the adipogenesis-related changes in the 2D and 3D cultured 3T3-L1 cells, including (1) the enlargement and softening of the 3D spheroids, (2) a dramatic enhancement in lipid staining and the expression of adipogenesis-related genes, and (3) a decrease in mitochondrial and glycolytic metabolic function. By adding ROCK-i to the BIM-A, most of these BIM-A-induced effects were cancelled. The collective findings reported herein suggest that ROCK-i eliminated the PG-induced suppression of adipogenesis in the 3T3-L1 cells, accompanied by the formation of enlarged 3D spheroids. Such effects of adding ROCK-i to a PG in preadipocytes on cellular properties appear to be associated with the suppression of PG-induced adverse effects, and provide additional insight into our understanding of lipid-related research.
Keywords: 3-dimensional (3D) tissue culture; 3T3-L1 cell; ROCK; ROCK inhibitor; Rho-kinase; upper eyelid sulcus (DUES) deepening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Addition of ROCK inhibitors to prostaglandin derivative (PG) synergistically affects adipogenesis of the 3D spheroids of human orbital fibroblasts (HOFs).Hum Cell. 2022 Jan;35(1):125-132. doi: 10.1007/s13577-021-00623-y. Epub 2021 Sep 30. Hum Cell. 2022. PMID: 34591280
-
Simultaneous Use of ROCK Inhibitors and EP2 Agonists Induces Unexpected Effects on Adipogenesis and the Physical Properties of 3T3-L1 Preadipocytes.Int J Mol Sci. 2021 Apr 28;22(9):4648. doi: 10.3390/ijms22094648. Int J Mol Sci. 2021. PMID: 33925005 Free PMC article.
-
Addition of EP2 agonists to an FP agonist additively and synergistically modulates adipogenesis and the physical properties of 3D 3T3-L1 sphenoids.Prostaglandins Leukot Essent Fatty Acids. 2021 Aug;171:102315. doi: 10.1016/j.plefa.2021.102315. Epub 2021 Jul 3. Prostaglandins Leukot Essent Fatty Acids. 2021. PMID: 34246925
-
Unexpected Crosslinking Effects of a Human Thyroid Stimulating Monoclonal Autoantibody, M22, with IGF1 on Adipogenesis in 3T3L-1 Cells.Int J Mol Sci. 2023 Jan 6;24(2):1110. doi: 10.3390/ijms24021110. Int J Mol Sci. 2023. PMID: 36674625 Free PMC article.
-
Brimonidine Modulates the ROCK1 Signaling Effects on Adipogenic Differentiation in 2D and 3D 3T3-L1 Cells.Bioengineering (Basel). 2022 Jul 19;9(7):327. doi: 10.3390/bioengineering9070327. Bioengineering (Basel). 2022. PMID: 35877378 Free PMC article.
Cited by
-
Unraveling White Adipose Tissue Heterogeneity and Obesity by Adipose Stem/Stromal Cell Biology and 3D Culture Models.Cells. 2023 Jun 8;12(12):1583. doi: 10.3390/cells12121583. Cells. 2023. PMID: 37371053 Free PMC article. Review.
-
From Eye Care to Hair Growth: Bimatoprost.Pharmaceuticals (Basel). 2024 Apr 27;17(5):561. doi: 10.3390/ph17050561. Pharmaceuticals (Basel). 2024. PMID: 38794131 Free PMC article. Review.
-
TGF-β effects on adipogenesis of 3T3-L1 cells differ in 2D and 3D cell culture conditions.FEBS Open Bio. 2024 Dec;14(12):2026-2037. doi: 10.1002/2211-5463.13890. Epub 2024 Oct 8. FEBS Open Bio. 2024. PMID: 39380256 Free PMC article.
-
Application of Single Cell Type-Derived Spheroids Generated by Using a Hanging Drop Culture Technique in Various In Vitro Disease Models: A Narrow Review.Cells. 2024 Sep 14;13(18):1549. doi: 10.3390/cells13181549. Cells. 2024. PMID: 39329734 Free PMC article. Review.
-
Rock inhibitors in Alzheimer's disease.Front Aging. 2025 Mar 20;6:1547883. doi: 10.3389/fragi.2025.1547883. eCollection 2025. Front Aging. 2025. PMID: 40182055 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

