A Broad-Host-Range Plasmid Outbreak: Dynamics of IncL/M Plasmids Transferring Carbapenemase Genes
- PMID: 36421285
- PMCID: PMC9686884
- DOI: 10.3390/antibiotics11111641
A Broad-Host-Range Plasmid Outbreak: Dynamics of IncL/M Plasmids Transferring Carbapenemase Genes
Abstract
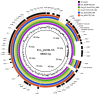
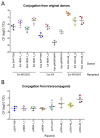
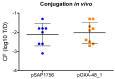
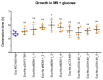
IncL/M broad-host-range conjugative plasmids are involved in the global spread of blaOXA-48 and the emergence of blaNDM-1. The aim of this study was to evaluate the transmission potential of plasmids encoding the emergent NDM-1 carbapenemase compared to the pandemic OXA-48. The conjugation rate and fitness cost of IncM2 and IncL plasmids encoding these carbapenemase genes were tested using a variety of host bacteria. Genomic analysis of uropathogenic Escherichia coli SAP1756 revealed that blaNDM-1 was encoded on an IncM2 plasmid, which also harboured blaTEM-1, bleMBL and sul1 and was highly similar to plasmids isolated from the same geographical area. Conjugation experiments demonstrated that NDM-1 and OXA-48-carrying plasmids transfer successfully between different Enterobacterales species, both in vitro and in vivo. Interestingly, E. coli isolates tested as recipients belonging to phylogroups A, B1, D and F were able to receive IncM2 plasmid pSAP1756, while phylogroups B2, C, E and G were not permissive to its acquisition. In general, the IncL OXA-48-carrying plasmids tested transferred at higher rates than IncM2 harbouring NDM-1 and imposed a lower burden to their host, possibly due to the inactivation of the tir fertility inhibition gene and reflecting their worldwide dissemination. IncM2 plasmids carrying blaNDM-1 are considered emergent threats that need continuous monitoring. In addition to sequencing efforts, phenotypic analysis of conjugation rates and fitness cost are effective methods for estimating the pandemic potential of antimicrobial resistance plasmids.
Keywords: Enterobacterales; IncL/M; NDM-1; OXA-48; antimicrobial resistance; carbapenemase; conjugation; plasmid.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- WHO . WHO publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. WHO; Geneva, Switzerland: 2017.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

