Octenidine's Efficacy: A Matter of Interpretation or the Influence of Experimental Setups?
- PMID: 36421309
- PMCID: PMC9686575
- DOI: 10.3390/antibiotics11111665
Octenidine's Efficacy: A Matter of Interpretation or the Influence of Experimental Setups?
Abstract
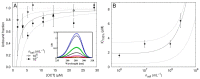
With its broad antimicrobial spectrum and non-specific mode of action via membrane disruption, any resistance to octenidine (OCT) seems unlikely and has not been observed in clinical settings so far. In this study, we aimed to investigate the efficacy of OCT against Escherichia coli and mutants lacking specific lipid head groups which, due to altered membrane properties, might be the root cause for resistance development of membrane-active compounds. Furthermore, we aimed to test its efficacy under different experimental conditions including different solvents for OCT, bacterial concentration and methods for analysis. Our primary goal was to estimate how many OCT molecules are needed to kill one bacterium. We performed susceptibility assays by observing bacterial growth behavior, using a Bioscreen in an analogous manner for every condition. The growth curves were recorded for 20 h at 420-580 nm in presence of different OCT concentrations and were used to assess the inhibitory concentrations (IC100%) for OCT. Bacterial concentrations given in cell numbers were determined, followed by Bioscreen measurement by manual colony counting on agar plates and QUANTOMTM cell staining. This indicated a significant variance between both methods, which influenced IC100% of OCT, especially when used at low doses. The binding capacity of OCT to E. coli was investigated by measuring UV-absorbance of OCT exposed to bacteria and a common thermodynamic framework based on Bioscreen measurements. Results showed that OCT's antimicrobial activity in E. coli is not affected by changes at the membrane level but strongly dependent on experimental settings in respect to solvents and applied bacterial counts. More OCT was required when the active was dissolved in phosphate or Hepes buffers instead of water and when higher bacterial concentration was used. Furthermore, binding studies revealed that 107-108 OCT molecules bind to bacteria, which is necessary for the saturation of the bacterial surface to initiate the killing cascade. Our results clearly demonstrate that in vitro data, depending on the applied materials and the methods for determination of IC100%, can easily be misinterpreted as reduced bacterial susceptibility towards OCT.
Keywords: Bioscreen; E. coli; MIC; QUANTOMTM; adaption; binding to bacteria; cell density; inoculum effect; lipid mutants; mechanism of action; multidrug resistance; octenidine; reduced susceptibility; resistance; tolerance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

