GOProFormer: A Multi-Modal Transformer Method for Gene Ontology Protein Function Prediction
- PMID: 36421723
- PMCID: PMC9687818
- DOI: 10.3390/biom12111709
GOProFormer: A Multi-Modal Transformer Method for Gene Ontology Protein Function Prediction
Abstract
Protein Language Models (PLMs) are shown to be capable of learning sequence representations useful for various prediction tasks, from subcellular localization, evolutionary relationships, family membership, and more. They have yet to be demonstrated useful for protein function prediction. In particular, the problem of automatic annotation of proteins under the Gene Ontology (GO) framework remains open. This paper makes two key contributions. It debuts a novel method that leverages the transformer architecture in two ways. A sequence transformer encodes protein sequences in a task-agnostic feature space. A graph transformer learns a representation of GO terms while respecting their hierarchical relationships. The learned sequence and GO terms representations are combined and utilized for multi-label classification, with the labels corresponding to GO terms. The method is shown superior over recent representative GO prediction methods. The second major contribution in this paper is a deep investigation of different ways of constructing training and testing datasets. The paper shows that existing approaches under- or over-estimate the generalization power of a model. A novel approach is proposed to address these issues, resulting in a new benchmark dataset to rigorously evaluate and compare methods and advance the state-of-the-art.
Keywords: gene ontology; multi-modal transformer; protein function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

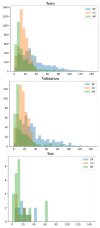
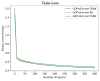

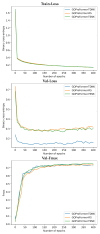

References
-
- Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser L., Polosukhin I. Attention Is All You Need. arXiv. 20171706.03762
-
- Elnaggar A., Heinzinger M., Dallago C., Rihawi G., Wang Y., Jones L., Gibbs T., Feher T., Angerer C., Steinegger M., et al. ProtTrans: Towards Cracking the Language of Lifes Code Through Self-Supervised Deep Learning and High Performance Computing. IEEE Trans. Patern Anal. Mach. Intell. 2021;44:7112–7127. doi: 10.1109/TPAMI.2021.3095381. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

