Analysis of the Genetic Variation of the Fruitless Gene within the Anopheles gambiae (Diptera: Culicidae) Complex Populations in Africa
- PMID: 36421951
- PMCID: PMC9699577
- DOI: 10.3390/insects13111048
Analysis of the Genetic Variation of the Fruitless Gene within the Anopheles gambiae (Diptera: Culicidae) Complex Populations in Africa
Abstract
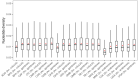
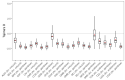
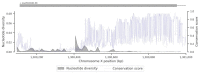
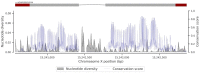
Targeting genes involved in sexual determinism, for vector or pest control purposes, requires a better understanding of their polymorphism in natural populations in order to ensure a rapid spread of the construct. By using genomic data from An. gambiae s.l., we analyzed the genetic variation and the conservation score of the fru gene in 18 natural populations across Africa. A total of 34,339 SNPs were identified, including 3.11% non-synonymous segregating sites. Overall, the nucleotide diversity was low, and the Tajima’s D neutrality test was negative, indicating an excess of low frequency SNPs in the fru gene. The allelic frequencies of the non-synonymous SNPs were low (freq < 0.26), except for two SNPs identified at high frequencies (freq > 0.8) in the zinc-finger A and B protein domains. The conservation score was variable throughout the fru gene, with maximum values in the exonic regions compared to the intronic regions. These results showed a low genetic variation overall in the exonic regions, especially the male sex-specific exon and the BTB-exon 1 of the fru gene. These findings will facilitate the development of an effective gene drive construct targeting the fru gene that can rapidly spread without encountering resistance in wild populations.
Keywords: Africa; An. gambiae s.l; Fruitless; genomics; vector control.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures











Similar articles
-
Genomic analyses revealed low genetic variation in the intron-exon boundary of the doublesex gene within the natural populations of An. gambiae s.l. in Burkina Faso.BMC Genomics. 2024 Dec 18;25(1):1207. doi: 10.1186/s12864-024-11127-y. BMC Genomics. 2024. PMID: 39695373 Free PMC article.
-
Genetic analysis and population structure of the Anopheles gambiae complex from different ecological zones of Burkina Faso.Infect Genet Evol. 2020 Jul;81:104261. doi: 10.1016/j.meegid.2020.104261. Epub 2020 Feb 22. Infect Genet Evol. 2020. PMID: 32092481
-
Genomic organisation of the neural sex determination gene fruitless (fru) in the Hawaiian species Drosophila silvestris and the conservation of the fru BTB protein-protein-binding domain throughout evolution.Hereditas. 2000;132(1):67-78. doi: 10.1111/j.1601-5223.2000.00067.x. Hereditas. 2000. PMID: 10857262
-
Insecticide resistance in Bemisia tabaci Gennadius (Homoptera: Aleyrodidae) and Anopheles gambiae Giles (Diptera: Culicidae) could compromise the sustainability of malaria vector control strategies in West Africa.Acta Trop. 2013 Oct;128(1):7-17. doi: 10.1016/j.actatropica.2013.06.004. Epub 2013 Jun 17. Acta Trop. 2013. PMID: 23792227 Review.
-
Genetic dissection of sexual orientation: behavioral, cellular, and molecular approaches in Drosophila melanogaster.Neurosci Res. 1996 Oct;26(2):95-107. doi: 10.1016/s0168-0102(96)01087-5. Neurosci Res. 1996. PMID: 8953572 Review.
Cited by
-
Symbiotic Wolbachia in mosquitoes and its role in reducing the transmission of mosquito-borne diseases: updates and prospects.Front Microbiol. 2023 Oct 13;14:1267832. doi: 10.3389/fmicb.2023.1267832. eCollection 2023. Front Microbiol. 2023. PMID: 37901801 Free PMC article. Review.
-
Genomic analyses revealed low genetic variation in the intron-exon boundary of the doublesex gene within the natural populations of An. gambiae s.l. in Burkina Faso.BMC Genomics. 2024 Dec 18;25(1):1207. doi: 10.1186/s12864-024-11127-y. BMC Genomics. 2024. PMID: 39695373 Free PMC article.
References
-
- Salvemini M., Mauro U., Lombardo F., Milano A., Zazzaro V., Arcà B., Polito L.C., Saccone G. Genomic organization and splicing evolution of the Doublesex gene, a Drosophila regulator of sexual differentiation, in the dengue and yellow fever mosquito Aedes aegypti. BMC Evol. Biol. 2011;11:41. doi: 10.1186/1471-2148-11-41. - DOI - PMC - PubMed
-
- Biedler J.K., Tu Z. Advances in Insect Physiology. Elsevier; Amsterdam, The Netherlands: 2016. Sex determination in mosquitoes; pp. 37–66.
LinkOut - more resources
Full Text Sources

