Developing New Tools to Fight Human Pathogens: A Journey through the Advances in RNA Technologies
- PMID: 36422373
- PMCID: PMC9697208
- DOI: 10.3390/microorganisms10112303
Developing New Tools to Fight Human Pathogens: A Journey through the Advances in RNA Technologies
Abstract
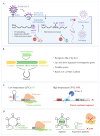
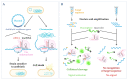
A long scientific journey has led to prominent technological advances in the RNA field, and several new types of molecules have been discovered, from non-coding RNAs (ncRNAs) to riboswitches, small interfering RNAs (siRNAs) and CRISPR systems. Such findings, together with the recognition of the advantages of RNA in terms of its functional performance, have attracted the attention of synthetic biologists to create potent RNA-based tools for biotechnological and medical applications. In this review, we have gathered the knowledge on the connection between RNA metabolism and pathogenesis in Gram-positive and Gram-negative bacteria. We further discuss how RNA techniques have contributed to the building of this knowledge and the development of new tools in synthetic biology for the diagnosis and treatment of diseases caused by pathogenic microorganisms. Infectious diseases are still a world-leading cause of death and morbidity, and RNA-based therapeutics have arisen as an alternative way to achieve success. There are still obstacles to overcome in its application, but much progress has been made in a fast and effective manner, paving the way for the solid establishment of RNA-based therapies in the future.
Keywords: CRISPR; RNA chaperones; RNA metabolism; RNA regulators; RNA tool; ribonucleases; small non-coding RNAs (ncRNAs); synthetic biology; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Arraiano C.M., Andrade J.M., Domingues S., Guinote I.B., Malecki M., Matos R.G., Moreira R.N., Pobre V., Reis F.P., Saramago M., et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

