Immunopeptidomic Analysis of BoLA-I and BoLA-DR Presented Peptides from Theileria parva Infected Cells
- PMID: 36423003
- PMCID: PMC9699068
- DOI: 10.3390/vaccines10111907
Immunopeptidomic Analysis of BoLA-I and BoLA-DR Presented Peptides from Theileria parva Infected Cells
Abstract
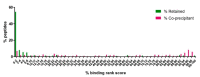
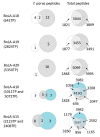
The apicomplexan parasite Theileria parva is the causative agent of East Coast fever, usually a fatal disease for cattle, which is prevalent in large areas of eastern, central, and southern Africa. Protective immunity against T. parva is mediated by CD8+ T cells, with CD4+ T-cells thought to be important in facilitating the full maturation and development of the CD8+ T-cell response. T. parva has a large proteome, with >4000 protein-coding genes, making T-cell antigen identification using conventional screening approaches laborious and expensive. To date, only a limited number of T-cell antigens have been described. Novel approaches for identifying candidate antigens for T. parva are required to replace and/or complement those currently employed. In this study, we report on the use of immunopeptidomics to study the repertoire of T. parva peptides presented by both BoLA-I and BoLA-DR molecules on infected cells. The study reports on peptides identified from the analysis of 13 BoLA-I and 6 BoLA-DR datasets covering a range of different BoLA genotypes. This represents the most comprehensive immunopeptidomic dataset available for any eukaryotic pathogen to date. Examination of the immunopeptidome data suggested the presence of a large number of coprecipitated and non-MHC-binding peptides. As part of the work, a pipeline to curate the datasets to remove these peptides was developed and used to generate a final list of 74 BoLA-I and 15 BoLA-DR-presented peptides. Together, the data demonstrated the utility of immunopeptidomics as a method to identify novel T-cell antigens for T. parva and the importance of careful curation and the application of high-quality immunoinformatics to parse the data generated.
Keywords: MHC; antigen processing and presentation; cattle; parasite-protozoan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Integral Use of Immunopeptidomics and Immunoinformatics for the Characterization of Antigen Presentation and Rational Identification of BoLA-DR-Presented Peptides and Epitopes.J Immunol. 2021 May 15;206(10):2489-2497. doi: 10.4049/jimmunol.2001409. Epub 2021 Mar 31. J Immunol. 2021. PMID: 33789985 Free PMC article.
-
Immunity and vaccine development in the bovine theilerioses.Adv Parasitol. 1999;44:41-97. doi: 10.1016/s0065-308x(08)60230-4. Adv Parasitol. 1999. PMID: 10563395 Review.
-
Use of "one-pot, mix-and-read" peptide-MHC class I tetramers and predictive algorithms to improve detection of cytotoxic T lymphocyte responses in cattle.Vet Res. 2014 Apr 28;45(1):50. doi: 10.1186/1297-9716-45-50. Vet Res. 2014. PMID: 24775445 Free PMC article.
-
Designing bovine T cell vaccines via reverse immunology.Ticks Tick Borne Dis. 2012 Jun;3(3):188-92. doi: 10.1016/j.ttbdis.2011.12.001. Epub 2012 Jan 9. Ticks Tick Borne Dis. 2012. PMID: 22621863 Review.
-
Limited diversity in the CD8+ antigen-coding loci in Theileria parva parasites from cattle from southern and eastern Africa.Vet Parasitol. 2021 Mar;291:109371. doi: 10.1016/j.vetpar.2021.109371. Epub 2021 Feb 5. Vet Parasitol. 2021. PMID: 33621717
Cited by
-
An immunoinformatics study reveals a new BoLA-DR-restricted CD4+ T cell epitopes on the Gag protein of bovine leukemia virus.Sci Rep. 2023 Dec 15;13(1):22356. doi: 10.1038/s41598-023-48899-4. Sci Rep. 2023. PMID: 38102157 Free PMC article.
-
Immunopeptidomic MHC-I profiling and immunogenicity testing identifies Tcj2 as a new Chagas disease mRNA vaccine candidate.PLoS Pathog. 2024 Dec 18;20(12):e1012764. doi: 10.1371/journal.ppat.1012764. eCollection 2024 Dec. PLoS Pathog. 2024. PMID: 39693359 Free PMC article.
References
-
- East Coast Fever. [(accessed on 30 September 2022)]. Available online: https://www.galvmed.org/livestock-and-diseases/livestock-diseases/east-c...
-
- Goddeeris B.M., Morrison W.I., Teale A.J. Generation of bovine cytotoxic cell lines, specific for cells infected with the protozoan parasite Theileria parva and restricted by products of the major histocompatibility complex. Eur. J. Immunol. 1986;16:1243–1249. doi: 10.1002/eji.1830161010. - DOI - PubMed
-
- Taracha E.L., Goddeeris B., Teale A.J., Kemp S.J., Morrison W.I. Parasite strain specificity of bovine cytotoxic T cell responses to Theileria parva is determined primarily by immunodominance. J. Immunol. 1995;155:4854–4860. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

