Phosphatidylinositol 4,5-Bisphosphate Mediates the Co-Distribution of Influenza A Hemagglutinin and Matrix Protein M1 at the Plasma Membrane
- PMID: 36423118
- PMCID: PMC9698905
- DOI: 10.3390/v14112509
Phosphatidylinositol 4,5-Bisphosphate Mediates the Co-Distribution of Influenza A Hemagglutinin and Matrix Protein M1 at the Plasma Membrane
Abstract
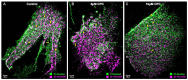
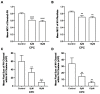
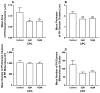
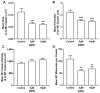
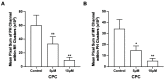
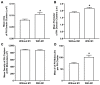
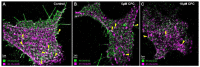
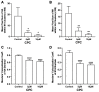
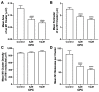
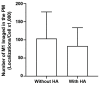
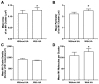
The fully assembled influenza A virus (IAV) has on its surface the highest density of a single membrane protein found in nature-the glycoprotein hemagglutinin (HA) that mediates viral binding, entry, and assembly. HA clusters at the plasma membrane of infected cells, and the HA density (number of molecules per unit area) of these clusters correlates with the infectivity of the virus. Dense HA clusters are considered to mark the assembly site and ultimately lead to the budding of infectious IAV. The mechanism of spontaneous HA clustering, which occurs with or without other viral components, has not been elucidated. Using super-resolution fluorescence photoactivation localization microscopy (FPALM), we have previously shown that these HA clusters are interdependent on phosphatidylinositol 4,5-biphosphate (PIP2). Here, we show that the IAV matrix protein M1 co-clusters with PIP2, visualized using the pleckstrin homology domain. We find that cetylpyridinium chloride (CPC), which is a positively charged quaternary ammonium compound known for its antibacterial and antiviral properties at millimolar concentrations, disrupts M1 clustering and M1-PIP2 co-clustering at micromolar concentrations well below the critical micelle concentration (CMC). CPC also disrupts the co-clustering of M1 with HA at the plasma membrane, suggesting the role of host cell PIP2 clusters as scaffolds for gathering and concentrating M1 and HA to achieve their unusually high cluster densities in the IAV envelope.
Keywords: FPALM; PIP2; cetylpyridinium chloride; hemagglutinin; host–virus interactions; influenza A; matrix protein M1; phosphoinositides; super-resolution fluorescence microscopy; virus assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The Effects of Viral Structural Proteins on Acidic Phospholipids in Host Membranes.Viruses. 2024 Oct 31;16(11):1714. doi: 10.3390/v16111714. Viruses. 2024. PMID: 39599829 Free PMC article. Review.
-
Cetylpyridinium chloride (CPC) reduces zebrafish mortality from influenza infection: Super-resolution microscopy reveals CPC interference with multiple protein interactions with phosphatidylinositol 4,5-bisphosphate in immune function.Toxicol Appl Pharmacol. 2022 Apr 1;440:115913. doi: 10.1016/j.taap.2022.115913. Epub 2022 Feb 9. Toxicol Appl Pharmacol. 2022. PMID: 35149080 Free PMC article.
-
A Defect in Influenza A Virus Particle Assembly Specific to Primary Human Macrophages.mBio. 2018 Oct 23;9(5):e01916-18. doi: 10.1128/mBio.01916-18. mBio. 2018. PMID: 30352935 Free PMC article.
-
Palmitoylation Contributes to Membrane Curvature in Influenza A Virus Assembly and Hemagglutinin-Mediated Membrane Fusion.J Virol. 2017 Oct 13;91(21):e00947-17. doi: 10.1128/JVI.00947-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28794042 Free PMC article.
-
Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses.Virus Res. 2001 Sep;77(1):61-9. doi: 10.1016/s0168-1702(01)00266-0. Virus Res. 2001. PMID: 11451488 Review.
Cited by
-
Conserved sequence features in intracellular domains of viral spike proteins.Virology. 2024 Nov;599:110198. doi: 10.1016/j.virol.2024.110198. Epub 2024 Aug 2. Virology. 2024. PMID: 39116647
-
Single influenza A viruses induce nanoscale cellular reprogramming at the virus-cell interface.Nat Commun. 2025 Apr 25;16(1):3846. doi: 10.1038/s41467-025-58935-8. Nat Commun. 2025. PMID: 40280912 Free PMC article.
-
PIP2 promotes the incorporation of CD43, PSGL-1 and CD44 into nascent HIV-1 particles.bioRxiv [Preprint]. 2024 Sep 5:2024.09.05.611432. doi: 10.1101/2024.09.05.611432. bioRxiv. 2024. Update in: Sci Adv. 2025 Apr 04;11(14):eads9711. doi: 10.1126/sciadv.ads9711. PMID: 39282414 Free PMC article. Updated. Preprint.
-
The bat influenza A virus subtype H18N11 induces nanoscale MHCII clustering upon host cell attachment.Nat Commun. 2025 Apr 25;16(1):3847. doi: 10.1038/s41467-025-58834-y. Nat Commun. 2025. PMID: 40280957 Free PMC article.
-
The Effects of Viral Structural Proteins on Acidic Phospholipids in Host Membranes.Viruses. 2024 Oct 31;16(11):1714. doi: 10.3390/v16111714. Viruses. 2024. PMID: 39599829 Free PMC article. Review.
References
-
- Gudheti M.V., Curthoys N.M., Gould T.J., Kim D., Gunewardene M.S., Gabor K.A., Gosse J.A., Kim C.H., Zimmerberg J., Hess S.T. Actin Mediates the Nanoscale Membrane Organization of the Clustered Membrane Protein Influenza Hemagglutinin. Biophys. J. 2013;104:2182–2192. doi: 10.1016/j.bpj.2013.03.054. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

