Fine Mapping the Soybean Mosaic Virus Resistance Gene in Soybean Cultivar Heinong 84 and Development of CAPS Markers for Rapid Identification
- PMID: 36423142
- PMCID: PMC9697120
- DOI: 10.3390/v14112533
Fine Mapping the Soybean Mosaic Virus Resistance Gene in Soybean Cultivar Heinong 84 and Development of CAPS Markers for Rapid Identification
Abstract
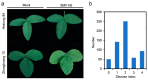
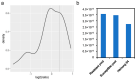
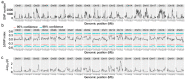
Heinong 84 is one of the major soybean varieties growing in Northeast China, and is resistant to the infection of all strains of soybean mosaic virus (SMV) in the region including the most prevalent strain, N3. However, the resistance gene(s) in Heinong 84 and the resistant mechanism are still elusive. In this study, genetic and next-generation sequencing (NGS)-based bulk segregation analysis (BSA) were performed to map the resistance gene using a segregation population from the cross of Heinong 84 and a susceptible cultivar to strain N3, Zhonghuang 13. Results show that the resistance of Heinong 84 is controlled by a dominant gene on chromosome 13. Further analyses suggest that the resistance gene in Heinong 84 is probably an allele of Rsv1. Finally, two pairs of single-nucleotide-polymorphism (SNP)-based primers that are tightly cosegregated with the resistance gene were designed for rapidly identifying resistant progenies in breeding via the cleaved amplified polymorphic sequence (CAPS) assay.
Keywords: Rsv1; bulk segregation analysis (BSA); cleaved amplified polymorphic sequences (CAPS) markers; resistance locus; soybean mosaic virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hill J.H., Whitham S.A. Advances in Virus Research. Elsevier; Amsterdam, The Netherlands: 2014. pp. 355–390. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

