Identification of Three Viruses Infecting Mulberry Varieties
- PMID: 36423172
- PMCID: PMC9696721
- DOI: 10.3390/v14112564
Identification of Three Viruses Infecting Mulberry Varieties
Abstract
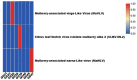
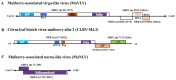
Viruses-mediated genome editing in plants is a powerful strategy to develop plant cultivars with important and novel agricultural traits. Mulberry alba is an important economic tree species that has been cultivated in China for more than 5000 years. So far, only a few viruses have been identified from mulberry trees, and their application potential is largely unknown. Therefore, mining more virus resources from the mulberry tree can pave the way for the establishment of useful engineering tools. In this study, eight old mulberry plants were gathered in seven geographic areas for virome analysis. Based on transcriptome analysis, we discovered three viruses associated with mulberries: Citrus leaf blotch virus isolate mulberry alba 2 (CLBV-ML2), Mulberry-associated virga-like virus (MaVLV), and Mulberry-associated narna-like virus (MaNLV). The genome of CLBV-ML2 was completely sequenced and exhibited high homology with Citriviruses, considered to be members of the genus Citrivirus, while the genomes of MaVLV and MaNLV were nearly completed lacking the 5' and 3' termini sequences. We tentatively consider MaVLV to be members of the family Virgaviridae and MaNLV to be members of the genus Narnavirus based on the results of phylogenetic trees. The infection experiments showed that CLBV-ML2 could be detected in the inoculated seedlings of both N. benthamiana and Morus alba, while MaVLV could only be detected in N. benthamiana. All of the infected seedlings did not show obvious symptoms.
Keywords: Citrivirus; Narnavirus; Virgaviridae; mulberry alba; transcriptome; virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Kwon S., Bodaghi S., Dang T., Gadhave K., Ho T., Osman F., Rwahnih M.A., Tzanetakis I., Simon A., Vidalakis G. Complete Nucleotide Sequence, Genome Organization, and Comparative Genomic Analyses of Citrus Yellow-Vein Associated Virus (CYVaV) Front. Microbiol. 2021;12:683130. doi: 10.3389/fmicb.2021.683130. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

