Intranasal pediatric parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in monkeys
- PMID: 36423629
- PMCID: PMC9684001
- DOI: 10.1016/j.cell.2022.11.006
Intranasal pediatric parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in monkeys
Abstract
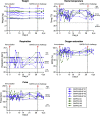
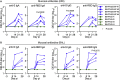
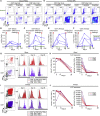
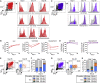
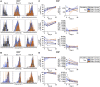
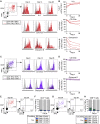
Pediatric SARS-CoV-2 vaccines are needed that elicit immunity directly in the airways as well as systemically. Building on pediatric parainfluenza virus vaccines in clinical development, we generated a live-attenuated parainfluenza-virus-vectored vaccine candidate expressing SARS-CoV-2 prefusion-stabilized spike (S) protein (B/HPIV3/S-6P) and evaluated its immunogenicity and protective efficacy in rhesus macaques. A single intranasal/intratracheal dose of B/HPIV3/S-6P induced strong S-specific airway mucosal immunoglobulin A (IgA) and IgG responses. High levels of S-specific antibodies were also induced in serum, which efficiently neutralized SARS-CoV-2 variants of concern of alpha, beta, and delta lineages, while their ability to neutralize Omicron sub-lineages was lower. Furthermore, B/HPIV3/S-6P induced robust systemic and pulmonary S-specific CD4+ and CD8+ T cell responses, including tissue-resident memory cells in the lungs. Following challenge, SARS-CoV-2 replication was undetectable in airways and lung tissues of immunized macaques. B/HPIV3/S-6P will be evaluated clinically as pediatric intranasal SARS-CoV-2/parainfluenza virus type 3 vaccine.
Keywords: SARS-CoV-2; intranasal vaccine; live-attenuated viral vector; mucosal immmunization; nasal spray vaccine; next-generation COVID vaccine; parainfluenza virus; pediatric; pediatric COVID vaccine.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests X.L., C.L., C.L.N., S.M., and U.J.B. are inventors on the provisional patent application number 63/180,534, entitled “recombinant chimeric B/HPIV3 expressing SARS-CoV-2 spike protein and its use,” filed by the United States Department of Health and Human Services.
Figures




Similar articles
-
Intranasal pediatric parainfluenza virus-vectored SARS-CoV-2 vaccine candidate is protective in macaques.bioRxiv [Preprint]. 2022 May 23:2022.05.21.492923. doi: 10.1101/2022.05.21.492923. bioRxiv. 2022. Update in: Proc Natl Acad Sci U S A. 2021 Dec 14;118(50):e2109744118. doi: 10.1073/pnas.2109744118. PMID: 35665011 Free PMC article. Updated. Preprint.
-
Intranasal parainfluenza virus-vectored vaccine expressing SARS-CoV-2 spike protein of Delta or Omicron B.1.1.529 induces mucosal and systemic immunity and protects hamsters against homologous and heterologous challenge.PLoS Pathog. 2025 Apr 21;21(4):e1012585. doi: 10.1371/journal.ppat.1012585. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40258004 Free PMC article.
-
Live-attenuated pediatric parainfluenza vaccine expressing 6P-stabilized SARS-CoV-2 spike protein is protective against SARS-CoV-2 variants in hamsters.bioRxiv [Preprint]. 2022 Dec 13:2022.12.12.520032. doi: 10.1101/2022.12.12.520032. bioRxiv. 2022. Update in: PLoS Pathog. 2023 Jun 23;19(6):e1011057. doi: 10.1371/journal.ppat.1011057. PMID: 36561185 Free PMC article. Updated. Preprint.
-
Live-attenuated pediatric parainfluenza vaccine expressing 6P-stabilized SARS-CoV-2 spike protein is protective against SARS-CoV-2 variants in hamsters.PLoS Pathog. 2023 Jun 23;19(6):e1011057. doi: 10.1371/journal.ppat.1011057. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37352333 Free PMC article.
-
A single intranasal dose of a live-attenuated parainfluenza virus-vectored SARS-CoV-2 vaccine is protective in hamsters.Proc Natl Acad Sci U S A. 2021 Dec 14;118(50):e2109744118. doi: 10.1073/pnas.2109744118. Proc Natl Acad Sci U S A. 2021. PMID: 34876520 Free PMC article.
Cited by
-
COVID-19 Vaccines for Optimizing Immunity in the Upper Respiratory Tract.Viruses. 2023 Oct 31;15(11):2203. doi: 10.3390/v15112203. Viruses. 2023. PMID: 38005881 Free PMC article. Review.
-
Mucosal immune response in biology, disease prevention and treatment.Signal Transduct Target Ther. 2025 Jan 8;10(1):7. doi: 10.1038/s41392-024-02043-4. Signal Transduct Target Ther. 2025. PMID: 39774607 Free PMC article.
-
Molecular Mechanism of VSV-Vectored ASFV Vaccine Activating Immune Response in DCs.Vet Sci. 2025 Jan 9;12(1):36. doi: 10.3390/vetsci12010036. Vet Sci. 2025. PMID: 39852910 Free PMC article.
-
A parainfluenza virus 5 (PIV5)-vectored intranasal SARS-CoV-2 vaccine (CVXGA1) elicits protective and long-lasting immunity in nonhuman primates.J Virol. 2025 Apr 15;99(4):e0199024. doi: 10.1128/jvi.01990-24. Epub 2025 Mar 21. J Virol. 2025. PMID: 40116506 Free PMC article.
-
Mucosal prime-boost immunization with live murine pneumonia virus-vectored SARS-CoV-2 vaccine is protective in macaques.Nat Commun. 2024 Apr 26;15(1):3553. doi: 10.1038/s41467-024-47784-6. Nat Commun. 2024. PMID: 38670948 Free PMC article.
References
-
- Funk A.L., Florin T.A., Kuppermann N., Tancredi D.J., Xie J., Kim K., Neuman M.I., Ambroggio L., Plint A.C., Mintegi S., et al. Outcomes of SARS-CoV-2-positive youths tested in emergency departments: the global PERN-COVID-19 study. JAMA Netw. Open. 2022;5:e2142322. doi: 10.1001/jamanetworkopen.2021.42322. - DOI - PMC - PubMed
-
- Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., Bonanomi E., D'Antiga L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

