Protein aggregation and calcium dysregulation are hallmarks of familial Parkinson's disease in midbrain dopaminergic neurons
- PMID: 36424392
- PMCID: PMC9691718
- DOI: 10.1038/s41531-022-00423-7
Protein aggregation and calcium dysregulation are hallmarks of familial Parkinson's disease in midbrain dopaminergic neurons
Abstract
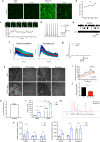
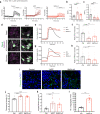
Mutations in the SNCA gene cause autosomal dominant Parkinson's disease (PD), with loss of dopaminergic neurons in the substantia nigra, and aggregation of α-synuclein. The sequence of molecular events that proceed from an SNCA mutation during development, to end-stage pathology is unknown. Utilising human-induced pluripotent stem cells (hiPSCs), we resolved the temporal sequence of SNCA-induced pathophysiological events in order to discover early, and likely causative, events. Our small molecule-based protocol generates highly enriched midbrain dopaminergic (mDA) neurons: molecular identity was confirmed using single-cell RNA sequencing and proteomics, and functional identity was established through dopamine synthesis, and measures of electrophysiological activity. At the earliest stage of differentiation, prior to maturation to mDA neurons, we demonstrate the formation of small β-sheet-rich oligomeric aggregates, in SNCA-mutant cultures. Aggregation persists and progresses, ultimately resulting in the accumulation of phosphorylated α-synuclein aggregates. Impaired intracellular calcium signalling, increased basal calcium, and impairments in mitochondrial calcium handling occurred early at day 34-41 post differentiation. Once midbrain identity fully developed, at day 48-62 post differentiation, SNCA-mutant neurons exhibited mitochondrial dysfunction, oxidative stress, lysosomal swelling and increased autophagy. Ultimately these multiple cellular stresses lead to abnormal excitability, altered neuronal activity, and cell death. Our differentiation paradigm generates an efficient model for studying disease mechanisms in PD and highlights that protein misfolding to generate intraneuronal oligomers is one of the earliest critical events driving disease in human neurons, rather than a late-stage hallmark of the disease.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Grants and funding
- MR/S006591/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_17179/MRC_/Medical Research Council/United Kingdom
- NC/X001067/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- 212251/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_00018/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

