Enrichment of antibiotic resistance genes within bacteriophage populations in saliva samples from individuals undergoing oral antibiotic treatments
- PMID: 36425042
- PMCID: PMC9678940
- DOI: 10.3389/fmicb.2022.1049110
Enrichment of antibiotic resistance genes within bacteriophage populations in saliva samples from individuals undergoing oral antibiotic treatments
Abstract
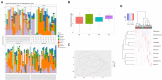
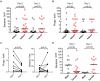
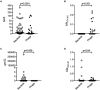
Spread of antibiotic resistance is a significant challenge for our modern health care system, and even more so in developing countries with higher prevalence of both infections and resistant bacteria. Faulty usage of antibiotics has been pinpointed as a driving factor in spread of resistant bacteria through selective pressure. However, horizontal gene transfer mediated through bacteriophages may also play an important role in this spread. In a cohort of Tanzanian patients suffering from bacterial infections, we demonstrate significant differences in the oral microbial diversity between infected and non-infected individuals, as well as before and after oral antibiotics treatment. Further, the resistome carried both by bacteria and bacteriophages vary significantly, with bla CTX-M1 resistance genes being mobilized and enriched within phage populations. This may impact how we consider spread of resistance in a biological context, as well in terms of treatment regimes.
Keywords: Bacteriophage; antibiotic resistance; antibiotic treatment; microbiota; saliva.
Copyright © 2022 Andersson, Makenga, Francis, Minja, Overballe-Petersen, Tang, Fuursted, Baraka and Lood.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Andersson T., Adell A. D., Moreno-Switt A. I., Spégel P., Turner C., Overballe-Petersen S., et al. (2022). Biogeographical variation in antimicrobial resistance in rivers is influenced by agriculture and is spread through bacteriophages. Environ. Microbiol. doi: 10.1111/1462-2920.16122 [Epub ahead of print], PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

