Molecular understanding of calorimetric protein unfolding experiments
- PMID: 36425081
- PMCID: PMC9680786
- DOI: 10.1016/j.bpr.2021.100037
Molecular understanding of calorimetric protein unfolding experiments
Abstract
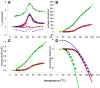
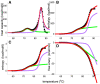
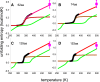
Testing and predicting protein stability gained importance because proteins, including antibodies, became pharmacologically relevant in viral and cancer therapies. Isothermal scanning calorimetry is the principle method to study protein stability. Here, we use the excellent experimental heat capacity Cp(T) data from the literature for a critical inspection of protein unfolding as well as for the test of a new cooperative model. In the relevant literature, experimental temperature profiles of enthalpy, Hcal(T), entropy, Scal(T), and free energy, Gcal(T) are missing. First, we therefore calculate the experimental Hcal(T), Scal(T), and Gcal(T) from published Cp(T) thermograms. Considering only the unfolding transition proper, the heat capacity and all thermodynamic functions are zero in the region of the native protein. In particular, the free energy of the folded proteins is also zero and Gcal(T) displays a trapezoidal temperature profile when cold denaturation is included. Second, we simulate the DSC-measured thermodynamic properties with a new molecular model based on statistical-mechanical thermodynamics. The model quantifies the protein cooperativity and predicts the aggregate thermodynamic variables of the system with molecular parameters only. The new model provides a perfect simulation of all thermodynamic properties, including the observed trapezoidal Gcal(T) temperature profile. Importantly, the new cooperative model can be applied to a broad range of protein sizes, including antibodies. It predicts not only heat and cold denaturation but also provides estimates of the unfolding kinetics and allows a comparison with molecular dynamics calculations and quasielastic neutron scattering experiments.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Murphy K.P., Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv. Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. https://www.ncbi.nlm.nih.gov/pubmed/1442323 - DOI - PubMed
-
- Privalov P.L., Gill S.J. Stability of protein-structure and hydrophobic interaction. Adv. Protein Chem. 1988;39:191–234. - PubMed
-
- Ibarra-Molero B., Sanchez-Ruiz J.M. Statistical differential scanning calorimetry: probing protein folding-unfolding ensembles. RSC Biomol. Sci. 2008:85–105. doi: 10.1039/9781847558282-00085. - DOI
-
- Brandts J.F. Thermodynamics of protein denaturation. I. Denaturation of chymotrypsinogen. J. Am. Chem. Soc. 1964;86:4291. doi: 10.1021/ja01074a013. - DOI
LinkOut - more resources
Full Text Sources

