Hsc70-4 aggravates PolyQ-mediated neurodegeneration by modulating NF-κB mediated immune response in Drosophila
- PMID: 36425218
- PMCID: PMC9678916
- DOI: 10.3389/fnmol.2022.857257
Hsc70-4 aggravates PolyQ-mediated neurodegeneration by modulating NF-κB mediated immune response in Drosophila
Abstract
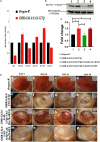
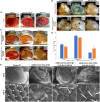
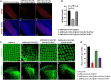
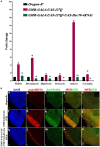
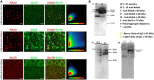
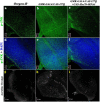
Huntington's disease occurs when the stretch of CAG repeats in exon 1 of the huntingtin (htt) gene crosses the permissible limit, causing the mutated protein (mHtt) to form insoluble aggregates or inclusion bodies. These aggregates are non-typically associated with various essential proteins in the cells, thus disrupting cellular homeostasis. The cells try to bring back normalcy by synthesizing evolutionary conserved cellular chaperones, and Hsp70 is one of the families of heat shock proteins that has a significant part in this, which comprises of heat-inducible and cognate forms. Here, we demonstrate that the heat shock cognate (Hsc70) isoform, Hsc70-4/HSPA8, has a distinct role in polyglutamate (PolyQ)-mediated pathogenicity, and its expression is enhanced in the polyQ conditions in Drosophila. Downregulation of hsc70-4 rescues PolyQ pathogenicity with a notable improvement in the ommatidia arrangement and near-normal restoration of optic neurons leading to improvement in phototaxis response. Reduced hsc70-4 also attenuates the augmented immune response by decreasing the expression of NF-κB and the antimicrobial peptides, along with that JNK overactivation is also restored. These lead to the rescue of the photoreceptor cells, indicating a decrease in the caspase activity, thus reverting the PolyQ pathogenicity. At the molecular level, we show the interaction between Hsc70-4, Polyglutamine aggregates, and NF-κB, which may be responsible for the dysregulation of signaling molecules in polyQ conditions. Thus, the present data provides a functional link between Hsc70-4 and NF-κB under polyQ conditions.
Keywords: Chaperone; HSPA8; Hsc70-4; Immune response; NF-κB; Neurodegeneration; PolyQ; Relish.
Copyright © 2022 Rai and Tapadia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Heat shock promotes inclusion body formation of mutant huntingtin (mHtt) and alleviates mHtt-induced transcription factor dysfunction.J Biol Chem. 2018 Oct 5;293(40):15581-15593. doi: 10.1074/jbc.RA118.002933. Epub 2018 Aug 24. J Biol Chem. 2018. PMID: 30143534 Free PMC article.
-
Molecular interaction between the chaperone Hsc70 and the N-terminal flank of huntingtin exon 1 modulates aggregation.J Biol Chem. 2015 Jan 30;290(5):2560-76. doi: 10.1074/jbc.M114.603332. Epub 2014 Dec 10. J Biol Chem. 2015. PMID: 25505179 Free PMC article.
-
Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity.Hum Mol Genet. 2000 Aug 12;9(13):2009-18. doi: 10.1093/hmg/9.13.2009. Hum Mol Genet. 2000. PMID: 10942430
-
Protein Misfolding and Aggregation as a Therapeutic Target for Polyglutamine Diseases.Brain Sci. 2017 Oct 11;7(10):128. doi: 10.3390/brainsci7100128. Brain Sci. 2017. PMID: 29019918 Free PMC article. Review.
-
[Molecular therapy targeting protein misfolding and aggregation for the polyglutamine diseases].Rinsho Shinkeigaku. 2009 Nov;49(11):913-6. doi: 10.5692/clinicalneurol.49.913. Rinsho Shinkeigaku. 2009. PMID: 20030247 Review. Japanese.
Cited by
-
Insights into dentatorubral-pallidoluysian atrophy from a new Drosophila model of disease.Neurobiol Dis. 2025 Apr;207:106834. doi: 10.1016/j.nbd.2025.106834. Epub 2025 Feb 5. Neurobiol Dis. 2025. PMID: 39921111 Free PMC article.
-
Closest horizons of Hsp70 engagement to manage neurodegeneration.Front Mol Neurosci. 2023 Sep 19;16:1230436. doi: 10.3389/fnmol.2023.1230436. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37795273 Free PMC article. Review.
-
Protein Misfolding in Pregnancy: Current Insights, Potential Mechanisms, and Implications for the Pathogenesis of Preeclampsia.Molecules. 2024 Jan 26;29(3):610. doi: 10.3390/molecules29030610. Molecules. 2024. PMID: 38338354 Free PMC article. Review.
References
-
- Arya R., Lakhotia S. C. (2006). A simple nail polish imprint technique for examination of external morphology of Drosophila eyes. Curr. Sci. 90, 1179–1180.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

