Bioinformatic analysis identifies HPV-related tumor microenvironment remodeling prognostic biomarkers in head and neck squamous cell carcinoma
- PMID: 36425786
- PMCID: PMC9679011
- DOI: 10.3389/fcimb.2022.1007950
Bioinformatic analysis identifies HPV-related tumor microenvironment remodeling prognostic biomarkers in head and neck squamous cell carcinoma
Abstract
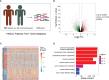
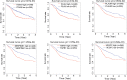
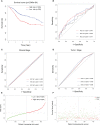
Head and neck squamous cell carcinomas (HNSCCs) are highly aggressive tumors with rapid progression and poor prognosis. Human papillomavirus (HPV) infection has been identified as one of the most important carcinogens for HNSCC. As an early event in HNSCC, infection with HPV leads to altered immune profiles in the tumor microenvironment (TME). The TME plays a key role in the progression and transformation of HNSCC. However, the TME in HNSCC is a complex and heterogeneous mix of tumor cells, fibroblasts, different types of infiltrating immune cells, and extracellular matrix. Biomarkers relevant to the TME, and the biological role of these biomarkers, remain poorly understood. To this end, we performed comprehensive analysis of the RNA sequencing (RNA-Seq) data from tumor tissue of 502 patients with HNSCC and healthy tissue of 44 control samples. In total, we identified 4,237 differentially expressed genes, including 2,062 upregulated and 2,175 downregulated genes. Further in-depth bioinformatic analysis suggested 19 HNSCC tumor tissue-specific genes. In the subsequent analysis, we focused on the biomarker candidates shown to be significantly associated with unfavorable patient survival: ITGA5, PLAU, PLAUR, SERPINE1, TGFB1, and VEGFC. We found that the expression of these genes was negatively regulated by DNA methylation. Strikingly, all of these potential biomarkers are profoundly involved in the activation of the epithelial-mesenchymal transition (EMT) pathway in HNSCCs. In addition, these targets were found to be positively correlated with the immune invasion levels of CD4+ T cells, macrophages, neutrophils, and dendritic cells, but negatively correlated with B-cell infiltration and CD8+ T-cell invasion. Notably, our data showed that the expression levels of ITGA5, PLAU, PLAUR, SERPINE1, and TGFB1 were significantly overexpressed in HPV-positive HNSCCs compared to normal controls, indicating the potential role of these biomarkers as transformation and/or malignant progression markers for HNSCCs in patients with HPV infection. Taken together, the results of our study propose ITGA5, PLAU, PLAUR, SERPINE1, and TGFB1 as potential prognostic biomarkers for HNSCCs, which might be involved in the HPV-related TME remodeling of HNSCC. Our findings provide important implications for the development and/or improvement of patient stratification and customized immunotherapies in HNSCC.
Keywords: HPV; biomarkers; head and neck squamous cell carcinoma; immune evasion; immunotherapy; tumor microenvironment.
Copyright © 2022 Zhou, Yuan, Cui, Hu, Xiao, Wei, Zhang and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer WuZ declared a shared affiliation with authors QZ and OY, to the handling editor at the time of review.
Figures





References
-
- Bellone G., Aste-Amezaga M., Trinchieri G., Rodeck U. (1995). Regulation of NK cell functions by TGF-β1. J. Immunol. 155, 1066–1073. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

