Genome-scale reconstruction and metabolic modelling of the fast-growing thermophile Geobacillus sp. LC300
- PMID: 36425956
- PMCID: PMC9678985
- DOI: 10.1016/j.mec.2022.e00212
Genome-scale reconstruction and metabolic modelling of the fast-growing thermophile Geobacillus sp. LC300
Abstract
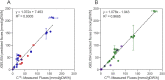
Thermophilic microorganisms show high potential for use as biorefinery cell factories. Their high growth temperatures provide fast conversion rates, lower risk of contaminations, and facilitated purification of volatile products. To date, only a few thermophilic species have been utilized for microbial production purposes, and the development of production strains is impeded by the lack of metabolic engineering tools. In this study, we constructed a genome-scale metabolic model, an important part of the metabolic engineering pipeline, of the fast-growing thermophile Geobacillus sp. LC300. The model (iGEL604) contains 604 genes, 1249 reactions and 1311 metabolites, and the reaction reversibility is based on thermodynamics at the optimum growth temperature. The growth phenotype is analyzed by batch cultivations on two carbon sources, further closing balances in carbon and degree-of-reduction. The predictive ability of the model is benchmarked against experimentally determined growth characteristics and internal flux distributions, showing high similarity to experimental phenotypes.
Keywords: Flux sampling; Genome-scale metabolic modelling; Geobacillus; Metabolic engineering; Thermophile.
© 2022 Published by Elsevier B.V. on behalf of International Metabolic Engineering Society.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

