The specific applications of the TSR-based method in identifying Zn2+ binding sites of proteases and ACE/ACE2
- PMID: 36426009
- PMCID: PMC9679521
- DOI: 10.1016/j.dib.2022.108629
The specific applications of the TSR-based method in identifying Zn2+ binding sites of proteases and ACE/ACE2
Abstract
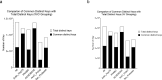
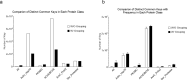
We have developed an alignment-free TSR (Triangular Spatial Relationship)-based computational method for protein structural comparison and motif identification and discovery. To demonstrate the potential applications of the method, we have generated two datasets. One dataset contains five classes: Actin/Hsp70, serine protease (chymotrypsin/trypsin/elastase), ArsC/Prdx2, PKA/PKB/PKC, and AChE/BChE at the hierarchical level 1 and twelve groups at the level 2. The other dataset includes representative proteases and ACE/ACE2. The x,y, z coordinates of the structures were obtained from PDB. We calculated the keys (or features) that represent each structure using the TSR-based method. The dataset and data presented here include additional information that help the readers become aware of specific applications of the TSR-based method in protein clustering, identification and discovery of metal ion binding sites as well as to understand the effect of amino acid grouping on protein 3D structural relationships at both global and local levels.
Keywords: 3D structure; Alignment-free; Amino acid grouping; Metal ion binding site; Protein similarity; Structural motif; Structure comparison; TSR.
Published by Elsevier Inc.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Figures











Similar articles
-
Introducing mirror-image discrimination capability to the TSR-based method for capturing stereo geometry and understanding hierarchical structure relationships of protein receptor family.Comput Biol Chem. 2023 Apr;103:107824. doi: 10.1016/j.compbiolchem.2023.107824. Epub 2023 Feb 3. Comput Biol Chem. 2023. PMID: 36753783 Free PMC article.
-
Development of the TSR-based computational method to investigate spike and monoclonal antibody interactions.Front Chem. 2025 Mar 19;13:1395374. doi: 10.3389/fchem.2025.1395374. eCollection 2025. Front Chem. 2025. PMID: 40177350 Free PMC article.
-
Exploring the effectiveness of the TSR-based protein 3-D structural comparison method for protein clustering, and structural motif identification and discovery of protein kinases, hydrolases, and SARS-CoV-2's protein via the application of amino acid grouping.Comput Biol Chem. 2021 Jun;92:107479. doi: 10.1016/j.compbiolchem.2021.107479. Epub 2021 Mar 29. Comput Biol Chem. 2021. PMID: 33951604
-
Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold.FEBS Lett. 1989 Jan 30;243(2):103-14. doi: 10.1016/0014-5793(89)80109-7. FEBS Lett. 1989. PMID: 2645167 Review.
-
Structure-function relationship of serine protease-protein inhibitor interaction.Acta Biochim Pol. 2001;48(2):419-28. Acta Biochim Pol. 2001. PMID: 11732612 Review.
Cited by
-
Introducing mirror-image discrimination capability to the TSR-based method for capturing stereo geometry and understanding hierarchical structure relationships of protein receptor family.Comput Biol Chem. 2023 Apr;103:107824. doi: 10.1016/j.compbiolchem.2023.107824. Epub 2023 Feb 3. Comput Biol Chem. 2023. PMID: 36753783 Free PMC article.
-
Development of the TSR-based computational method to investigate spike and monoclonal antibody interactions.Front Chem. 2025 Mar 19;13:1395374. doi: 10.3389/fchem.2025.1395374. eCollection 2025. Front Chem. 2025. PMID: 40177350 Free PMC article.
-
Development of a novel representation of drug 3D structures and enhancement of the TSR-based method for probing drug and target interactions.Comput Biol Chem. 2024 Oct;112:108117. doi: 10.1016/j.compbiolchem.2024.108117. Epub 2024 Jun 4. Comput Biol Chem. 2024. PMID: 38852360
References
-
- Kondra S, Chen F, Chen Y, Chen Y, Collette CJ, Xu W. A study of a hierarchical structure of proteins and ligand binding sites of receptors using the triangular spatial relationship-based structure comparison method and development of a size-filtering feature designed for comparing different sizes of protein structures. Proteins. 2022;90(1):239–257. doi: 10.1002/prot.26215. - DOI - PubMed
-
- Sarkar T, Raghavan VV, Chen F, Riley A, Zhou S, Xu W. Exploring the effectiveness of the TSR-based protein 3-D structural comparison method for protein clustering, and structural motif identification and discovery of protein kinases, hydrolase, and SARS-CoV-2’s protein via the application of amino acid grouping. Comput. Biol. Chem. 2021 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

