Chromosomal assembly of the flat oyster (Ostrea edulis L.) genome as a new genetic resource for aquaculture
- PMID: 36426129
- PMCID: PMC9679248
- DOI: 10.1111/eva.13462
Chromosomal assembly of the flat oyster (Ostrea edulis L.) genome as a new genetic resource for aquaculture
Abstract
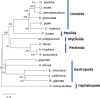
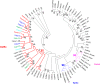
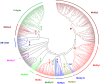
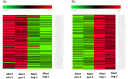
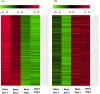
The European flat oyster (Ostrea edulis L.) is a native bivalve of the European coasts. Harvest of this species has declined during the last decades because of the appearance of two parasites that have led to the collapse of the stocks and the loss of the natural oyster beds. O. edulis has been the subject of numerous studies in population genetics and on the detection of the parasites Bonamia ostreae and Marteilia refringens. These studies investigated immune responses to these parasites at the molecular and cellular levels. Several genetic improvement programs have been initiated especially for parasite resistance. Within the framework of a European project (PERLE 2) that aims to produce genetic lines of O. edulis with hardiness traits (growth, survival, resistance) for the purpose of repopulating natural oyster beds in Brittany and reviving the culture of this species in the foreshore, obtaining a reference genome becomes essential as done recently in many bivalve species of aquaculture interest. Here, we present a chromosome-level genome assembly and annotation for the European flat oyster, generated by combining PacBio, Illumina, 10X linked, and Hi-C sequencing. The finished assembly is 887.2 Mb with a scaffold-N50 of 97.1 Mb scaffolded on the expected 10 pseudochromosomes. Annotation of the genome revealed the presence of 35,962 protein-coding genes. We analyzed in detail the transposable element (TE) diversity in the flat oyster genome, highlighted some specificities in tRNA and miRNA composition, and provided the first insight into the molecular response of O. edulis to M. refringens. This genome provides a reference for genomic studies on O. edulis to better understand its basic physiology and as a useful resource for genetic breeding in support of aquaculture and natural reef restoration.
Keywords: Martelia; aquaculture; flat oyster; genome; transposable elements.
© 2022 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Admire, A. , Shanks, L. , Danzl, N. , Wang, M. , Weier, U. , Stevens, W. , Hunt, E. , & Weinert, T. (2006). Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes and Development, 20(2), 159–173. 10.1101/gad.1392506 - DOI - PMC - PubMed
-
- Bai, C. M. , Xin, L. S. , Rosani, U. , Wu, B. , Wang, Q. C. , Duan, X. K. , Liu, Z. H. , & Wang, C. M. (2019). Chromosomal‐level assembly of the blood clam, Scapharca (Anadara) broughtonii, using long sequence reads and Hi‐C. Gigascience, 8(7), giz067. 10.1093/gigascience/giz067 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

