Dysregulation of HNF1B/Clusterin axis enhances disease progression in a highly aggressive subset of pancreatic cancer patients
- PMID: 36426859
- PMCID: PMC10122429
- DOI: 10.1093/carcin/bgac092
Dysregulation of HNF1B/Clusterin axis enhances disease progression in a highly aggressive subset of pancreatic cancer patients
Abstract
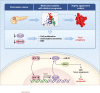
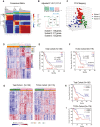
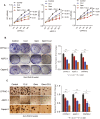
Pancreatic ductal adenocarcinoma (PDAC) is a lethal malignancy and is largely refractory to available treatments. Identifying key pathways associated with disease aggressiveness and therapeutic resistance may characterize candidate targets to improve patient outcomes. We used a strategy of examining the tumors from a subset of PDAC patient cohorts with the worst survival to understand the underlying mechanisms of aggressive disease progression and to identify candidate molecular targets with potential therapeutic significance. Non-negative matrix factorization (NMF) clustering, using gene expression profile, revealed three patient subsets. A 142-gene signature specific to the subset with the worst patient survival, predicted prognosis and stratified patients with significantly different survival in the test and validation cohorts. Gene-network and pathway analysis of the 142-gene signature revealed dysregulation of Clusterin (CLU) in the most aggressive patient subset in our patient cohort. Hepatocyte nuclear factor 1 b (HNF1B) positively regulated CLU, and a lower expression of HNF1B and CLU was associated with poor patient survival. Mechanistic and functional analyses revealed that CLU inhibits proliferation, 3D spheroid growth, invasiveness and epithelial-to-mesenchymal transition (EMT) in pancreatic cancer cell lines. Mechanistically, CLU enhanced proteasomal degradation of EMT-regulator, ZEB1. In addition, orthotopic transplant of CLU-expressing pancreatic cancer cells reduced tumor growth in mice. Furthermore, CLU enhanced sensitivity of pancreatic cancer cells representing aggressive patient subset, to the chemotherapeutic drug gemcitabine. Taken together, HNF1B/CLU axis negatively regulates pancreatic cancer progression and may potentially be useful in designing novel strategies to attenuate disease progression in PDAC patients.
Published by Oxford University Press 2022.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Siegel, R.L., et al. . (2022) Cancer statistics, 2022. CA Cancer J. Clin., 72, 7–33. - PubMed
-
- Rahib, L., et al. . (2014) Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res., 74, 29134006–29132921. - PubMed
-
- Bailey, P., et al. . (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature, 531, 47–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

