MicroRNAs are deeply linked to the emergence of the complex octopus brain
- PMID: 36427315
- PMCID: PMC9699675
- DOI: 10.1126/sciadv.add9938
MicroRNAs are deeply linked to the emergence of the complex octopus brain
Abstract
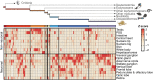
Soft-bodied cephalopods such as octopuses are exceptionally intelligent invertebrates with a highly complex nervous system that evolved independently from vertebrates. Because of elevated RNA editing in their nervous tissues, we hypothesized that RNA regulation may play a major role in the cognitive success of this group. We thus profiled messenger RNAs and small RNAs in three cephalopod species including 18 tissues of the Octopus vulgaris. We show that the major RNA innovation of soft-bodied cephalopods is an expansion of the microRNA (miRNA) gene repertoire. These evolutionarily novel miRNAs were primarily expressed in adult neuronal tissues and during the development and had conserved and thus likely functional target sites. The only comparable miRNA expansions happened, notably, in vertebrates. Thus, we propose that miRNAs are intimately linked to the evolution of complex animal brains.
Figures




References
-
- B. Hochner, T. Shomrat, G. Fiorito,The octopus: A model for a comparative analysis of the evolution of learning and memory mechanisms. Biol. Bull. 210,308–317 (2006). - PubMed
-
- J. Z. Young, Anatomy of the nervous system of Octopus vulgaris (1971); https://agris.fao.org/agris-search/search.do?recordID=US201300479148.
-
- W.-S. Chung, N. D. Kurniawan, N. J. Marshall,Comparative brain structure and visual processing in octopus from different habitats. Curr. Biol. 32,97–110.e4 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

