Revealing Genetic Diversity and Population Structure of Endangered Altay White-Headed Cattle Population Using 100 k SNP Markers
- PMID: 36428441
- PMCID: PMC9686749
- DOI: 10.3390/ani12223214
Revealing Genetic Diversity and Population Structure of Endangered Altay White-Headed Cattle Population Using 100 k SNP Markers
Abstract
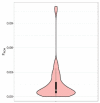
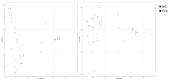
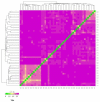
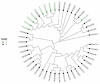
Understanding the genetic basis of native cattle populations that have adapted to the local environment is of great significance for formulating appropriate strategies and programs for genetic improvement and protection. Therefore, it is necessary to understand the genetic diversity and population structure of Altay white-headed cattle so as to meet the current production needs under various environments, carry out continuous genetic improvement, and promote rapid adaptation to changing environments and breeding objectives. A total of 46 individual samples of endangered Xinjiang Altay white-headed cattle were collected in this study, including nine bulls and 37 cows. To collect genotype data, 100 k SNP markers were used, and then studies of genetic diversity, genetic structure, inbreeding degree, and family analysis were carried out. A total of 101,220 SNP loci were detected, and the genotype detection rate for individuals was ≥90%. There were 85,993 SNP loci that passed quality control, of which 93.5% were polymorphic. The average effective allele number was 0.036, the Polymorphism Information Content was 0.304 and the minimum allele frequency was 0.309, the average observed heterozygosity was 0.413, and the average expected heterozygosity was 0.403. The average genetic distance of Idengtical By State (IBS) was 0.3090, there were 461 ROH (genome-length homozygous fragments), 76.1% of which were between 1 and 5 MB in length, and the average inbreeding coefficient was 0.016. The 46 Altay white-headed cattle were divided into their families, and the individual numbers of each family were obviously different. To sum up, the Altay white-headed cattle conservation population had low heterozygosity, a high inbreeding degree, few families, and large differences in the number of individuals in each family, which can easily cause a loss of genetic diversity. In the follow-up seed conservation process, seed selection and matching should be carried out according to the divided families to ensure the long-term protection of Altay white-headed cattle genetic resources.
Keywords: Altay white-headed cattle; SNP chip; genetic diversity; population structure; species in imminent danger.
Conflict of interest statement
All authors declare no conflict of interest.
Figures






Similar articles
-
Genome-wide analysis emancipates genomic diversity and signature of selection in Altay white-headed cattle of Xinjiang, China.Front Genet. 2023 Mar 30;14:1144249. doi: 10.3389/fgene.2023.1144249. eCollection 2023. Front Genet. 2023. PMID: 37065480 Free PMC article.
-
Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing.Yi Chuan. 2023 Jul 20;45(7):602-616. doi: 10.16288/j.yczz.23-115. Yi Chuan. 2023. PMID: 37503584
-
Analysis of the Genetic Relationship and Inbreeding Coefficient of the Hetian Qing Donkey through a Simplified Genome Sequencing Technology.Genes (Basel). 2024 Apr 28;15(5):570. doi: 10.3390/genes15050570. Genes (Basel). 2024. PMID: 38790199 Free PMC article.
-
Runs of homozygosity: current knowledge and applications in livestock.Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review.
-
Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits.BMC Genomics. 2021 Oct 15;22(1):747. doi: 10.1186/s12864-021-08042-x. BMC Genomics. 2021. PMID: 34654366 Free PMC article. Review.
Cited by
-
Assessing Genomic Diversity and Signatures of Selection in Chinese Red Steppe Cattle Using High-Density SNP Array.Animals (Basel). 2023 May 22;13(10):1717. doi: 10.3390/ani13101717. Animals (Basel). 2023. PMID: 37238146 Free PMC article.
-
Genome-wide analysis emancipates genomic diversity and signature of selection in Altay white-headed cattle of Xinjiang, China.Front Genet. 2023 Mar 30;14:1144249. doi: 10.3389/fgene.2023.1144249. eCollection 2023. Front Genet. 2023. PMID: 37065480 Free PMC article.
-
Genome-wide scan for SNPs and selective sweeps reveals candidate genes and QTLs for milk production and reproduction traits in Indian Kankrej cattle.3 Biotech. 2025 Apr;15(4):90. doi: 10.1007/s13205-025-04263-z. Epub 2025 Mar 14. 3 Biotech. 2025. PMID: 40092452
-
Development and Application of a 40 K Liquid Capture Chip for Beef Cattle.Animals (Basel). 2025 May 7;15(9):1346. doi: 10.3390/ani15091346. Animals (Basel). 2025. PMID: 40362161 Free PMC article.
-
Genetic Diversity and Selection Signatures of Lvliang Black Goat Using Genome-Wide SNP Data.Animals (Basel). 2024 Nov 3;14(21):3154. doi: 10.3390/ani14213154. Animals (Basel). 2024. PMID: 39518877 Free PMC article.
References
-
- Daka P. Introduction to Altay white-headeded Cattle. China’s Livest. Poult. Seed Ind. 2010;6:64–65.
-
- Yugang W., Xia L. Discussion on the Improvement of Livestock Varieties in Altay. Heilongjiang Anim. Breed. 2015;23:58–61.
-
- Hazi W.T., Muhan B., Bahati, Adali, Shaken, Hatipa Introduction to Altay white-headeded Cattle. Heilongjiang Anim. Husb. Vet. 2004;5:20–21.
-
- Hazi W.T., Sweat B.W. Altay white-headeded cattle, a local fine breed. Rural. Pract. Sci. Technol. Inf. 2006;11:23.
-
- Zhongrong Breeding conservation of Altay white-headeded cattle. Xinjiang Anim. Husb. 2015;4:41–43.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

