The Immunological Epigenetic Landscape of the Human Life Trajectory
- PMID: 36428462
- PMCID: PMC9687906
- DOI: 10.3390/biomedicines10112894
The Immunological Epigenetic Landscape of the Human Life Trajectory
Abstract
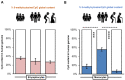
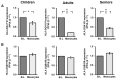
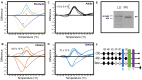
Adaptive immunity changes over an individual’s lifetime, maturing by adulthood and diminishing with old age. Epigenetic mechanisms involving DNA and histone methylation form the molecular basis of immunological memory during lymphocyte development. Monocytes alter their function to convey immune tolerance, yet the epigenetic influences at play remain to be fully understood in the context of lifespan. This study of a healthy genetically homogenous cohort of children, adults and seniors sought to decipher the epigenetic dynamics in B-lymphocytes and monocytes. Variable global cytosine methylation within retro-transposable LINE-1 repeats was noted in monocytes compared to B-lymphocytes across age groups. The expression of the human leukocyte antigen (HLA)-DQ alpha chain gene HLA-DQA1*01 revealed significantly reduced levels in monocytes in all ages relative to B-lymphocytes, as well as between lifespan groups. High melting point analysis and bisulfite sequencing of the HLA-DQA1*01 promoter in monocytes highlighted variable cytosine methylation in children and seniors but greater stability at this locus in adults. Further epigenetic evaluation revealed higher histone lysine 27 trimethylation in monocytes from this adult group. Chromatin immunoprecipitation and RNA pulldown demonstrated association with a novel lncRNA TINA with structurally conserved similarities to the previously recognized epigenetic modifier PARTICLE. Seeking to interpret the epigenetic immunological landscape across three representative age groups, this study focused on HLA-DQA1*01 to expose cytosine and histone methylation alterations and their association with the non-coding transcriptome. Such insights unveil previously unknown complex epigenetic layers, orchestrating the strength and weakening of adaptive immunity with the progression of life.
Keywords: ChIP; HLA; PARTICLE; TINA; histone; long non-coding RNA; methylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Differences in promoter DNA methylation and mRNA expression of individual alleles of the HLA class II DQA1 gene.Immunol Lett. 2015 Oct;167(2):147-54. doi: 10.1016/j.imlet.2015.08.006. Epub 2015 Aug 20. Immunol Lett. 2015. PMID: 26297837
-
Long non-coding RNA PARTICLE bridges histone and DNA methylation.Sci Rep. 2017 May 11;7(1):1790. doi: 10.1038/s41598-017-01875-1. Sci Rep. 2017. PMID: 28496150 Free PMC article.
-
DNA methylation and mRNA expression of HLA-DQA1 alleles in type 1 diabetes mellitus.Immunology. 2016 Jun;148(2):150-9. doi: 10.1111/imm.12593. Epub 2016 Mar 7. Immunology. 2016. PMID: 26854762 Free PMC article.
-
Epigenetic landscape of amphetamine and methamphetamine addiction in rodents.Epigenetics. 2015;10(7):574-80. doi: 10.1080/15592294.2015.1055441. Epigenetics. 2015. PMID: 26023847 Free PMC article. Review.
-
DNA Methylation and Immune Memory Response.Cells. 2021 Oct 29;10(11):2943. doi: 10.3390/cells10112943. Cells. 2021. PMID: 34831166 Free PMC article. Review.
References
-
- Laupeze B., Fardel O., Onno M., Bertho N., Drenou B., Fauchet R., Amiot L. Differential expression of major histocompatibility complex class Ia, Ib, and II molecules on monocytes-derived dendritic and macrophagic cells. Hum. Immunol. 1999;60:591–597. doi: 10.1016/S0198-8859(99)00025-7. - DOI - PubMed
-
- Krensky A.M. The HLA system, antigen processing and presentation. Kidney Int. Suppl. 1997;58:S2–S7. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

