Analysis of Gene Single Nucleotide Polymorphisms in COVID-19 Disease Highlighting the Susceptibility and the Severity towards the Infection
- PMID: 36428884
- PMCID: PMC9689844
- DOI: 10.3390/diagnostics12112824
Analysis of Gene Single Nucleotide Polymorphisms in COVID-19 Disease Highlighting the Susceptibility and the Severity towards the Infection
Abstract
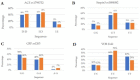
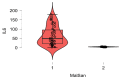
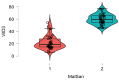
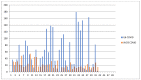
Many factors may influence the risk of being infected by SARS-CoV-2, the coronavirus responsible for coronavirus disease 2019 (COVID-19). Exposure to the virus cannot explain the variety of an individual's responses to the virus and the high differences of effect that the virus may cause to some. While a person's preexisting condition and their immune defenses have been confirmed to play a major role in the disease progression, there is still much to learn about hosts' genetic makeup towards COVID-19 susceptibility and risk. The host genetic makeup may have direct influence on the grade of predisposition and outcomes of COVID-19. In this study, we aimed to investigate the presence of relevant genetic single nucleotide polymorphisms (SNPs), the peripheral blood level of IL6, vitamin D and arterial blood gas (ABG) markers (pH, oxygen-SpO2 and carbon dioxide-SpCO2) on two groups, COVID-19 (n = 41, study), and the healthy (n = 43, control). We analyzed cytokine and interleukin genes in charge of both pro-inflammatory and immune-modulating responses and those genes that are considered involved in the COVID-19 progression and complications. Thus, we selected major genes, such as IL1β, IL1RN (IL-1 β and α receptor) IL6, IL6R (IL-6 receptor), IL10, IFNγ (interferon gamma), TNFα (tumor necrosis factor alpha), ACE2 (angiotensin converting enzyme), SERPINA3 (Alpha-1-Antiproteinase, Antitrypsin member of Serpin 3 family), VDR (vitamin D receptor Tak1, Bsm1 and Fok1), and CRP (c-reactive protein). Though more research is needed, these findings may give a better representation of virus pleiotropic activity and its relation to the immune system.
Keywords: COVID-19; SARS-CoV-2; arterial blood gas (ABG); carbon dioxide (SpCO2); oxygen (SpO2).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- SeyedAlinaghi S., Mehrtak M., MohsseniPour M., Mirzapour P., Barzegary A., Habibi P., Moradmand-Badie B., Afsahi A.M., Karimi A., Heydari M., et al. Genetic susceptibility of COVID-19: A systematic review of current evidence. Eur. J. Med. Res. 2021;26:46. doi: 10.1186/s40001-021-00516-8. - DOI - PMC - PubMed
-
- Driggin E., Madhavan M.V., Bikdeli B., Chuich T., Laracy J., Biondi-Zoccai G., Brown T.S., Der Nigoghossian C., Zidar D.A., Haythe J., et al. Cardiovascular Considerations for Patients, Health Care Workers, and Health Systems During the COVID-19 Pandemic. J. Am. Coll. Cardiol. 2020;75:2352–2371. doi: 10.1016/j.jacc.2020.03.031. - DOI - PMC - PubMed
-
- Balzanelli M.G., Distratis P., Catucci O., Amatulli F., Cefalo A., Lazzaro R., Aityan K.S., Dalagni G., Nico A., De Michele A., et al. Clinical and diagnostic findings in COVID-19 patients: An original research from SG Moscati Hospital in Taranto Italy. J. Biol. Regul. Homeost. Agents. 2021;35:171–183. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

