Episignature Mapping of TRIP12 Provides Functional Insight into Clark-Baraitser Syndrome
- PMID: 36430143
- PMCID: PMC9690904
- DOI: 10.3390/ijms232213664
Episignature Mapping of TRIP12 Provides Functional Insight into Clark-Baraitser Syndrome
Abstract
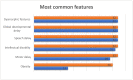
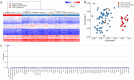
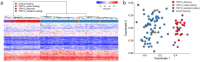
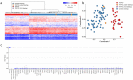
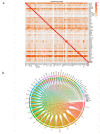
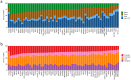
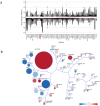
Clark-Baraitser syndrome is a rare autosomal dominant intellectual disability syndrome caused by pathogenic variants in the TRIP12 (Thyroid Hormone Receptor Interactor 12) gene. TRIP12 encodes an E3 ligase in the ubiquitin pathway. The ubiquitin pathway includes activating E1, conjugating E2 and ligating E3 enzymes which regulate the breakdown and sorting of proteins. This enzymatic pathway is crucial for physiological processes. A significant proportion of TRIP12 variants are currently classified as variants of unknown significance (VUS). Episignatures have been shown to represent a powerful diagnostic tool to resolve inconclusive genetic findings for Mendelian disorders and to re-classify VUSs. Here, we show the results of DNA methylation episignature analysis in 32 individuals with pathogenic, likely pathogenic and VUS variants in TRIP12. We identified a specific and sensitive DNA methylation (DNAm) episignature associated with pathogenic TRIP12 variants, establishing its utility as a clinical biomarker for Clark-Baraitser syndrome. In addition, we performed analysis of differentially methylated regions as well as functional correlation of the TRIP12 genome-wide methylation profile with the profiles of 56 additional neurodevelopmental disorders.
Keywords: Clark–Baraitser syndrome; DNA methylation; TRIP12; episignature; intellectual disability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bramswig N.C., Lüdecke H.-J., Pettersson M., Albrecht B., Bernier R.A., Cremer K., Eichler E.E., Falkenstein D., Gerdts J., Jansen S., et al. Identification of new TRIP12 variants and detailed clinical evaluation of individuals with non-syndromic intellectual disability with or without autism. Hum. Genet. 2017;136:179–192. doi: 10.1007/s00439-016-1743-x. - DOI - PMC - PubMed
-
- Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.-J., de Vries P., A de Vries B.B., Willemsen M.H., Kleefstra T., Löhner K., et al. Meta-analysis of 2104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

