The Role of the Exonic lncRNA PRKDC-210 in Transcription Regulation
- PMID: 36430260
- PMCID: PMC9692655
- DOI: 10.3390/ijms232213783
The Role of the Exonic lncRNA PRKDC-210 in Transcription Regulation
Abstract
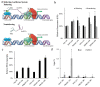
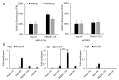
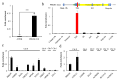
In recent years, long noncoding RNAs (lncRNAs) have received increasing attention and have been reported to be associated with various genetic abnormalities. However, the functions of many lncRNAs, including those of long exonic noncoding RNAs (lencRNAs), have not yet been elucidated. Here, we used a novel tethering luciferase assay to analyze the transcriptional regulatory functions of five lencRNAs that are upregulated in cancer. We found that the lencRNA PRKDC-210 interacts with MED12, a component of the CDK8 complex, to regulate the transcription of several genes. The transcriptional activation ability of PRKDC-210 was abolished in siRNA-treated CDK8-depleted cells. We also confirmed the enrichment of PRKDC-210 on RNA polymerase II. RNA-seq analysis of cells in which PRKDC-210 or PRKDC mRNA was knocked down using antisense oligonucleotides revealed that PRKDC-210 can affect the expression levels of genes related to fatty acid metabolism. Finally, we used a ChIRP assay to examine PRKDC-210-enriched sites in the genome. Overall, our findings demonstrate that the lencRNA PRKDC-210 promotes transcription through the CDK8 complex pathway at the transcription initiation site. We propose that PRKDC-210 can affect the transcription of adjacent genes after its transcription and splicing.
Keywords: CDK8; MED12; cancer; directed affect; lncRNA; tethering luciferase assay; transcription regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Divergent transcription is associated with promoters of transcriptional regulators.BMC Genomics. 2013 Dec 23;14:914. doi: 10.1186/1471-2164-14-914. BMC Genomics. 2013. PMID: 24365181 Free PMC article.
-
Long Noncoding RNA GMAN, Up-regulated in Gastric Cancer Tissues, Is Associated With Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS.Gastroenterology. 2019 Feb;156(3):676-691.e11. doi: 10.1053/j.gastro.2018.10.054. Epub 2018 Nov 13. Gastroenterology. 2019. PMID: 30445010
-
Expression of the Antisense-to-Latency Transcript Long Noncoding RNA in Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2017 Jan 31;91(4):e01698-16. doi: 10.1128/JVI.01698-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928018 Free PMC article.
-
Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases.Brain Res Bull. 2013 Aug;97:69-80. doi: 10.1016/j.brainresbull.2013.06.001. Epub 2013 Jun 10. Brain Res Bull. 2013. PMID: 23756188 Review.
-
Long noncoding RNAs: Novel insights into hepatocelluar carcinoma.Cancer Lett. 2014 Mar 1;344(1):20-27. doi: 10.1016/j.canlet.2013.10.021. Epub 2013 Oct 30. Cancer Lett. 2014. PMID: 24183851 Review.
Cited by
-
RNA Regulatory Networks 2.0.Int J Mol Sci. 2023 May 19;24(10):9001. doi: 10.3390/ijms24109001. Int J Mol Sci. 2023. PMID: 37240347 Free PMC article.
References
-
- Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J., et al. A Long Noncoding RNA LincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

