Deciphering the Role and Signaling Pathways of PKCα in Luminal A Breast Cancer Cells
- PMID: 36430510
- PMCID: PMC9696894
- DOI: 10.3390/ijms232214023
Deciphering the Role and Signaling Pathways of PKCα in Luminal A Breast Cancer Cells
Abstract
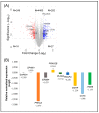
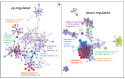
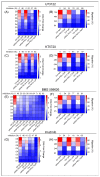
Protein kinase C (PKC) comprises a family of highly related serine/threonine protein kinases involved in multiple signaling pathways, which control cell proliferation, survival, and differentiation. The role of PKCα in cancer has been studied for many years. However, it has been impossible to establish whether PKCα acts as an oncogene or a tumor suppressor. Here, we analyzed the importance of PKCα in cellular processes such as proliferation, migration, or apoptosis by inhibiting its gene expression in a luminal A breast cancer cell line (MCF-7). Differential expression analysis and phospho-kinase arrays of PKCα-KD vs. PKCα-WT MCF-7 cells identified an essential set of proteins and oncogenic kinases of the JAK/STAT and PI3K/AKT pathways that were down-regulated, whereas IGF1R, ERK1/2, and p53 were up-regulated. In addition, unexpected genes related to the interferon pathway appeared down-regulated, while PLC, ERBB4, or PDGFA displayed up-regulated. The integration of this information clearly showed us the usefulness of inhibiting a multifunctional kinase-like PKCα in the first step to control the tumor phenotype. Then allowing us to design a possible selection of specific inhibitors for the unexpected up-regulated pathways to further provide a second step of treatment to inhibit the proliferation and migration of MCF-7 cells. The results of this study suggest that PKCα plays an oncogenic role in this type of breast cancer model. In addition, it reveals the signaling mode of PKCα at both gene expression and kinase activation. In this way, a wide range of proteins can implement a new strategy to fine-tune the control of crucial functions in these cells and pave the way for designing targeted cancer therapies.
Keywords: PKC; breast cancer; kinases; signaling pathways; targeted therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Mitotic checkpoint kinase Mps1/TTK predicts prognosis of colon cancer patients and regulates tumor proliferation and differentiation via PKCα/ERK1/2 and PI3K/Akt pathway.Med Oncol. 2019 Nov 13;37(1):5. doi: 10.1007/s12032-019-1320-y. Med Oncol. 2019. PMID: 31720873
-
Identification of a GαGβγ, AKT and PKCα signalome associated with invasive growth in two genetic models of human breast cancer cell epithelial-to-mesenchymal transition.Int J Oncol. 2012 Jul;41(1):189-200. doi: 10.3892/ijo.2012.1457. Epub 2012 Apr 30. Int J Oncol. 2012. PMID: 22552300
-
Protein kinase Calpha is involved in interferon regulatory factor 3 activation and type I interferon-beta synthesis.J Biol Chem. 2007 May 18;282(20):15022-32. doi: 10.1074/jbc.M700421200. Epub 2007 Feb 12. J Biol Chem. 2007. PMID: 17296604
-
The complexities of PKCα signaling in cancer.Adv Biol Regul. 2021 May;80:100769. doi: 10.1016/j.jbior.2020.100769. Epub 2020 Nov 23. Adv Biol Regul. 2021. PMID: 33307285 Free PMC article. Review.
-
Potential implications of protein kinase Cα in pathophysiological conditions and therapeutic interventions.Life Sci. 2023 Oct 1;330:121999. doi: 10.1016/j.lfs.2023.121999. Epub 2023 Aug 1. Life Sci. 2023. PMID: 37536614 Review.
Cited by
-
Zein-Derived Peptides from Corn Promote the Proliferation of C2C12 Myoblasts via Crosstalk of mTORC1 and mTORC2 Signaling Pathways.Foods. 2024 Mar 18;13(6):919. doi: 10.3390/foods13060919. Foods. 2024. PMID: 38540908 Free PMC article.
-
PKCα regulates the secretion of PDL1-carrying small extracellular vesicles in a p53-dependent manner.Cell Death Dis. 2025 Jan 14;16(1):19. doi: 10.1038/s41419-025-07341-5. Cell Death Dis. 2025. PMID: 39809736 Free PMC article.
-
Second-Generation Antipsychotics Induce Metabolic Disruption in Adipose Tissue-Derived Mesenchymal Stem Cells Through an aPKC-Dependent Pathway.Cells. 2024 Dec 17;13(24):2084. doi: 10.3390/cells13242084. Cells. 2024. PMID: 39768174 Free PMC article.
References
-
- Inoue M., Kishimoto A., Takai Y., Nishizuka Y. Studies on a Cyclic Nucleotide-Independent Protein Kinase and Its Proenzyme in Mammalian Tissues. II. Proenzyme and Its Activation by Calcium-Dependent Protease from Rat Brain. J. Biol. Chem. 1977;252:7610–7616. doi: 10.1016/S0021-9258(17)41010-6. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

