Novel CYP11A1-Derived Vitamin D and Lumisterol Biometabolites for the Management of COVID-19
- PMID: 36432468
- PMCID: PMC9698837
- DOI: 10.3390/nu14224779
Novel CYP11A1-Derived Vitamin D and Lumisterol Biometabolites for the Management of COVID-19
Abstract
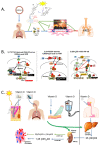
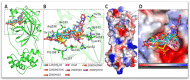
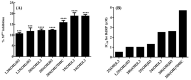
Vitamin D deficiency is associated with a higher risk of SARS-CoV-2 infection and poor outcomes of the COVID-19 disease. However, a satisfactory mechanism explaining the vitamin D protective effects is missing. Based on the anti-inflammatory and anti-oxidative properties of classical and novel (CYP11A1-derived) vitamin D and lumisterol hydroxymetabolites, we have proposed that they would attenuate the self-amplifying damage in lungs and other organs through mechanisms initiated by interactions with corresponding nuclear receptors. These include the VDR mediated inhibition of NFκβ, inverse agonism on RORγ and the inhibition of ROS through activation of NRF2-dependent pathways. In addition, the non-receptor mediated actions of vitamin D and related lumisterol hydroxymetabolites would include interactions with the active sites of SARS-CoV-2 transcription machinery enzymes (Mpro;main protease and RdRp;RNA dependent RNA polymerase). Furthermore, these metabolites could interfere with the binding of SARS-CoV-2 RBD with ACE2 by interacting with ACE2 and TMPRSS2. These interactions can cause the conformational and dynamical motion changes in TMPRSS2, which would affect TMPRSS2 to prime SARS-CoV-2 spike proteins. Therefore, novel, CYP11A1-derived, active forms of vitamin D and lumisterol can restrain COVID-19 through both nuclear receptor-dependent and independent mechanisms, which identify them as excellent candidates for antiviral drug research and for the educated use of their precursors as nutrients or supplements in the prevention and attenuation of the COVID-19 disease.
Keywords: ACE2; Mpro; RdRp; SARS-CoV-2; anti-inflammatory; lumisterol; vitamin D.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Vitamin D and lumisterol novel metabolites can inhibit SARS-CoV-2 replication machinery enzymes.Am J Physiol Endocrinol Metab. 2021 Aug 1;321(2):E246-E251. doi: 10.1152/ajpendo.00174.2021. Epub 2021 Jun 28. Am J Physiol Endocrinol Metab. 2021. PMID: 34181461 Free PMC article.
-
Photoprotective Properties of Vitamin D and Lumisterol Hydroxyderivatives.Cell Biochem Biophys. 2020 Jun;78(2):165-180. doi: 10.1007/s12013-020-00913-6. Epub 2020 May 22. Cell Biochem Biophys. 2020. PMID: 32441029 Free PMC article. Review.
-
Therapeutic potential of green tea catechin, (-)-epigallocatechin-3-O-gallate (EGCG) in SARS-CoV-2 infection: Major interactions with host/virus proteases.Phytomed Plus. 2023 Feb;3(1):100402. doi: 10.1016/j.phyplu.2022.100402. Epub 2022 Dec 30. Phytomed Plus. 2023. PMID: 36597465 Free PMC article. Review.
-
Withanone from Withania somnifera Attenuates SARS-CoV-2 RBD and Host ACE2 Interactions to Rescue Spike Protein Induced Pathologies in Humanized Zebrafish Model.Drug Des Devel Ther. 2021 Mar 11;15:1111-1133. doi: 10.2147/DDDT.S292805. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 33737804 Free PMC article.
-
Effects of vitamin C and D on the mRNA expression of angiotensin converting enzyme 2 receptor, cathepsin L, and transmembrane serine protease in the mouse lungs.Libyan J Med. 2022 Dec;17(1):2054111. doi: 10.1080/19932820.2022.2054111. Libyan J Med. 2022. PMID: 35311495 Free PMC article.
Cited by
-
Association of ApaI rs7975232 and BsmI rs1544410 in clinical outcomes of COVID-19 patients according to different SARS-CoV-2 variants.Sci Rep. 2023 Mar 3;13(1):3612. doi: 10.1038/s41598-023-30859-7. Sci Rep. 2023. PMID: 36869206 Free PMC article.
-
COVID-19: Focusing on the Link between Inflammation, Vitamin D, MAPK Pathway and Oxidative Stress Genetics.Antioxidants (Basel). 2023 May 20;12(5):1133. doi: 10.3390/antiox12051133. Antioxidants (Basel). 2023. PMID: 37237997 Free PMC article.
-
Recent Advances in Vitamin D Biology: Something New under the Sun.J Invest Dermatol. 2023 Dec;143(12):2340-2342. doi: 10.1016/j.jid.2023.07.003. Epub 2023 Oct 4. J Invest Dermatol. 2023. PMID: 37791933 Free PMC article. No abstract available.
-
Antioxidant Functions of Vitamin D and CYP11A1-Derived Vitamin D, Tachysterol, and Lumisterol Metabolites: Mechanisms, Clinical Implications, and Future Directions.Antioxidants (Basel). 2024 Aug 17;13(8):996. doi: 10.3390/antiox13080996. Antioxidants (Basel). 2024. PMID: 39199241 Free PMC article. Review.
-
Vitamin D3 and COVID-19 Outcomes: An Umbrella Review of Systematic Reviews and Meta-Analyses.Antioxidants (Basel). 2023 Jan 22;12(2):247. doi: 10.3390/antiox12020247. Antioxidants (Basel). 2023. PMID: 36829806 Free PMC article. Review.
References
-
- Holick M.F., Clark M.B. The photobiogenesis and metabolism of vitamin D. Fed. Proc. 1978;37:2567–2574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

