Gut Microbiota Associated with Gestational Health Conditions in a Sample of Mexican Women
- PMID: 36432504
- PMCID: PMC9696207
- DOI: 10.3390/nu14224818
Gut Microbiota Associated with Gestational Health Conditions in a Sample of Mexican Women
Abstract
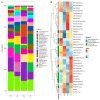
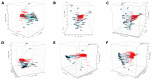
Gestational diabetes (GD), pre-gestational diabetes (PD), and pre-eclampsia (PE) are morbidities affecting gestational health which have been associated with dysbiosis of the mother's gut microbiota. This study aimed to assess the extent of change in the gut microbiota diversity, short-chain fatty acids (SCFA) production, and fecal metabolites profile in a sample of Mexican women affected by these disorders. Fecal samples were collected from women with GD, PD, or PE in the third trimester of pregnancy, along with clinical and biochemical data. Gut microbiota was characterized by high-throughput DNA sequencing of V3-16S rRNA gene libraries; SCFA and metabolites were measured by High-Pressure Liquid Chromatography (HPLC) and (Fourier Transform Ion Cyclotron Mass Spectrometry (FT-ICR MS), respectively, in extracts prepared from feces. Although the results for fecal microbiota did not show statistically significant differences in alfa diversity for GD, PD, and PE concerning controls, there was a difference in beta diversity for GD versus CO, and a high abundance of Proteobacteria, followed by Firmicutes and Bacteroidota among gestational health conditions. DESeq2 analysis revealed bacterial genera associated with each health condition; the Spearman's correlation analyses showed selected anthropometric, biochemical, dietary, and SCFA metadata associated with specific bacterial abundances, and although the HPLC did not show relevant differences in SCFA content among the studied groups, FT-ICR MS disclosed the presence of interesting metabolites of complex phenolic, valeric, arachidic, and caprylic acid nature. The major conclusion of our work is that GD, PD, and PE are associated with fecal bacterial microbiota profiles, with distinct predictive metagenomes.
Keywords: Bruker Solarix XR; FT ICR MS; fecal microbiota; gestational diabetes; gut microbiota; high-throughput DNA sequencing; mother feces; pre-eclampsia; pre-gestational diabetes.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
MeSH terms
Substances
Grants and funding
- SECTEI/249/2019-CM-SECTEI/109/2020-CM-SECTEI/124/2021, Convocatoria 2019, Proyectos Científicos, Tecnológicos y/o de Innovación para la atención a problemas específicos de la Ciudad de México relacionados con la investigación y atención de enfermedades cr/Secretaría de Ciencia, Tecnología e Innovación de la CDMX
- CONACyT-163235 INFR-2011-01/Consejo Nacional de Ciencia y Tecnología
- CONACyT-302670-2019-Apoyos para Adquisición y Mantenimiento de Infraestructura en Insti-tuciones y laboratorios de Investigación Especializada./Consejo Nacional de Ciencia y Tecnología
LinkOut - more resources
Full Text Sources

