A genome-wide association study identifies novel players in Na and Fe homeostasis in Arabidopsis thaliana under alkaline-salinity stress
- PMID: 36433704
- PMCID: PMC10108281
- DOI: 10.1111/tpj.16042
A genome-wide association study identifies novel players in Na and Fe homeostasis in Arabidopsis thaliana under alkaline-salinity stress
Abstract
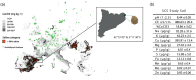
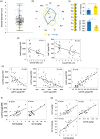
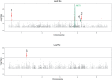
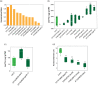
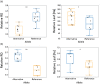
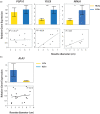
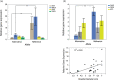
In nature, multiple stress factors occur simultaneously. The screening of natural diversity panels and subsequent Genome-Wide Association Studies (GWAS) is a powerful approach to identify genetic components of various stress responses. Here, the nutritional status variation of a set of 270 natural accessions of Arabidopsis thaliana grown on a natural saline-carbonated soil is evaluated. We report significant natural variation on leaf Na (LNa) and Fe (LFe) concentrations in the studied accessions. Allelic variation in the NINJA and YUC8 genes is associated with LNa diversity, and variation in the ALA3 is associated with LFe diversity. The allelic variation detected in these three genes leads to changes in their mRNA expression and correlates with plant differential growth performance when plants are exposed to alkaline salinity treatment under hydroponic conditions. We propose that YUC8 and NINJA expression patters regulate auxin and jasmonic signaling pathways affecting plant tolerance to alkaline salinity. Finally, we describe an impairment in growth and leaf Fe acquisition associated with differences in root expression of ALA3, encoding a phospholipid translocase active in plasma membrane and the trans Golgi network which directly interacts with proteins essential for the trafficking of PIN auxin transporters, reinforcing the role of phytohormonal processes in regulating ion homeostasis under alkaline salinity.
Keywords: Arabidopsis; GWAS; alkaline-salinity; ionome; natural variation.
© 2022 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene.J Exp Bot. 2016 Apr;67(8):2127-38. doi: 10.1093/jxb/erw015. Epub 2016 Feb 11. J Exp Bot. 2016. PMID: 26873976 Free PMC article.
-
The heterotrimeric G-protein β subunit, AGB1, plays multiple roles in the Arabidopsis salinity response.Plant Cell Environ. 2015 Oct;38(10):2143-56. doi: 10.1111/pce.12542. Epub 2015 May 12. Plant Cell Environ. 2015. PMID: 25808946
-
Fluctuating selection on migrant adaptive sodium transporter alleles in coastal Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):E12443-E12452. doi: 10.1073/pnas.1816964115. Epub 2018 Dec 7. Proc Natl Acad Sci U S A. 2018. PMID: 30530653 Free PMC article.
-
Ion homeostasis for salinity tolerance in plants: a molecular approach.Physiol Plant. 2021 Apr;171(4):578-594. doi: 10.1111/ppl.13185. Epub 2020 Aug 31. Physiol Plant. 2021. PMID: 32770745 Review.
-
A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K(+)/Na(+) ratio in leaves during salinity stress.Plant Cell Environ. 2010 Apr;33(4):552-65. doi: 10.1111/j.1365-3040.2009.02056.x. Epub 2009 Nov 4. Plant Cell Environ. 2010. PMID: 19895406 Review.
Cited by
-
Chasing the mechanisms of ecologically adaptive salinity tolerance.Plant Commun. 2023 Nov 13;4(6):100571. doi: 10.1016/j.xplc.2023.100571. Epub 2023 Mar 7. Plant Commun. 2023. PMID: 36883005 Free PMC article. Review.
References
-
- Alloway, B.J. (2013) Molybdenum. In: Heavy metals in soils. Dordrecht: Springer, pp. 527–534.
-
- Assefa, T. , Zhang, J. , Chowda‐Reddy, R.V. , Moran Lauter, A.N. , Singh, A. , O'Rourke, J.A. et al. (2020) Deconstructing the genetic architecture of iron deficiency chlorosis in soybean using genome‐wide approaches. BMC Plant Biology, 20(1), 1–13. Available from: 10.1186/s12870-020-2237-5 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

