A likely HOXC4 predisposition variant for Chiari malformations
- PMID: 36433874
- PMCID: PMC10193467
- DOI: 10.3171/2022.10.JNS22956
A likely HOXC4 predisposition variant for Chiari malformations
Abstract
Objective: Inherited variants predisposing patients to type 1 or 1.5 Chiari malformation (CM) have been hypothesized but have proven difficult to confirm. The authors used a unique high-risk pedigree population resource and approach to identify rare candidate variants that likely predispose individuals to CM and protein structure prediction tools to identify pathogenicity mechanisms.
Methods: By using the Utah Population Database, the authors identified pedigrees with significantly increased numbers of members with CM diagnosis. From a separate DNA biorepository of 451 samples from CM patients and families, 32 CM patients belonging to 1 or more of 24 high-risk Chiari pedigrees were identified. Two high-risk pedigrees had 3 CM-affected relatives, and 22 pedigrees had 2 CM-affected relatives. To identify rare candidate predisposition gene variants, whole-exome sequence data from these 32 CM patients belonging to 24 CM-affected related pairs from high-risk pedigrees were analyzed. The I-TASSER package for protein structure prediction was used to predict the structures of both the wild-type and mutant proteins found here.
Results: Sequence analysis of the 24 affected relative pairs identified 38 rare candidate Chiari predisposition gene variants that were shared by at least 1 CM-affected pair from a high-risk pedigree. The authors found a candidate variant in HOXC4 that was shared by 2 CM-affected patients in 2 independent pedigrees. All 4 of these CM cases, 2 in each pedigree, exhibited a specific craniocervical bony phenotype defined by a clivoaxial angle less than 125°. The protein structure prediction results suggested that the mutation considered here may reduce the binding affinity of HOXC4 to DNA.
Conclusions: Analysis of unique and powerful Utah genetic resources allowed identification of 38 strong candidate CM predisposition gene variants. These variants should be pursued in independent populations. One of the candidates, a rare HOXC4 variant, was identified in 2 high-risk CM pedigrees, with this variant possibly predisposing patients to a Chiari phenotype with craniocervical kyphosis.
Keywords: Chiari malformation; Utah Population Database; craniocervical kyphosis; high-risk pedigree; predisposition; protein prediction modeling.
Conflict of interest statement
The authors report no conflict of interest concerning the materials or methods used in this study or the findings specified in this paper.
Figures



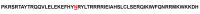
References
-
- Speer MC, George TM, Enterline DS, Franklin A, Wolpert CM, Milhorat TH. A genetic hypothesis for Chiari I malformation with or without syringomyelia. Neurosurg Focus. 2000;8(3):E12. - PubMed
-
- Boyles AL, Enterline DS, Hammock PH, et al. Phenotypic definition of Chiari type I malformation coupled with high-density SNP genome screen shows significant evidence for linkage to regions on chromosomes 9 and 15. Am J Med Genet A. 2006;140(24):2776–2785. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

