Translational readthrough of nonsense mutant TP53 by mRNA incorporation of 5-Fluorouridine
- PMID: 36433934
- PMCID: PMC9700717
- DOI: 10.1038/s41419-022-05431-2
Translational readthrough of nonsense mutant TP53 by mRNA incorporation of 5-Fluorouridine
Abstract
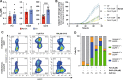
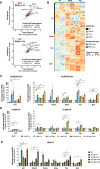
TP53 nonsense mutations in cancer produce truncated inactive p53 protein. We show that 5-FU metabolite 5-Fluorouridine (FUr) induces full-length p53 in human tumor cells carrying R213X nonsense mutant TP53. Ribosome profiling visualized translational readthrough at the R213X premature stop codon and demonstrated that FUr-induced readthrough is less permissive for canonical stop codon readthrough compared to aminoglycoside G418. FUr is incorporated into mRNA and can potentially base-pair with guanine, allowing insertion of Arg tRNA at the TP53 R213X UGA premature stop codon and translation of full-length wild-type p53. We confirmed that full-length p53 rescued by FUr triggers tumor cell death by apoptosis. FUr also restored full-length p53 in TP53 R213X mutant human tumor xenografts in vivo. Thus, we demonstrate a novel strategy for therapeutic rescue of nonsense mutant TP53 and suggest that FUr should be explored for treatment of patients with TP53 nonsense mutant tumors.
© 2022. The Author(s).
Conflict of interest statement
V.J.N.B. and K.G.W. are co-founders and shareholders of Aprea Therapeutics, a company that develops p53-based cancer therapy including APR-246. V.J.N.B. is employed by Aprea Therapeutics as data science consultant. K.G.W. is a member of its Clinical Advisory Board. Research in the K.G.W. lab has received financial support from Aprea Therapeutics. K.G.W. has received a salary from Aprea Therapeutics.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

