Restoring cellular magnesium balance through Cyclin M4 protects against acetaminophen-induced liver damage
- PMID: 36433951
- PMCID: PMC9700862
- DOI: 10.1038/s41467-022-34262-0
Restoring cellular magnesium balance through Cyclin M4 protects against acetaminophen-induced liver damage
Abstract
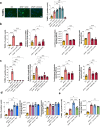
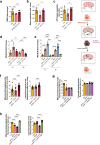
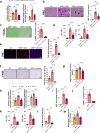
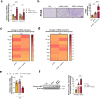
Acetaminophen overdose is one of the leading causes of acute liver failure and liver transplantation in the Western world. Magnesium is essential in several cellular processess. The Cyclin M family is involved in magnesium transport across cell membranes. Herein, we identify that among all magnesium transporters, only Cyclin M4 expression is upregulated in the liver of patients with acetaminophen overdose, with disturbances in magnesium serum levels. In the liver, acetaminophen interferes with the mitochondrial magnesium reservoir via Cyclin M4, affecting ATP production and reactive oxygen species generation, further boosting endoplasmic reticulum stress. Importantly, Cyclin M4 mutant T495I, which impairs magnesium flux, shows no effect. Finally, an accumulation of Cyclin M4 in endoplasmic reticulum is shown under hepatoxicity. Based on our studies in mice, silencing hepatic Cyclin M4 within the window of 6 to 24 h following acetaminophen overdose ingestion may represent a therapeutic target for acetaminophen overdose induced liver injury.
© 2022. The Author(s).
Conflict of interest statement
The rest of the authors declare no competing interest. U.S. is employed by Silence Therapeutics GmbH. This study is under the patent “Nucleic acids for inhibiting expression of Cnnm4 in a cell”. WO2021239825A1. Inventor: Ute Schaeper, Sibylle Dames, Steffen Schubert, Alfonso Martínez de la Cruz, Jorge Simón Espinosa, Irene González Recio, María Luz Martínez Chantar. (02/12/2021). The present patent previously described identifies the use of Galnac siRNA Cnnm4 for the treatment of liver disease. This article reinforces the validation of this therapeutic approach for the pathology of DILI due to paracetamol overdose.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

