GPSM1 impairs metabolic homeostasis by controlling a pro-inflammatory pathway in macrophages
- PMID: 36434066
- PMCID: PMC9700814
- DOI: 10.1038/s41467-022-34998-9
GPSM1 impairs metabolic homeostasis by controlling a pro-inflammatory pathway in macrophages
Abstract
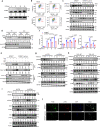
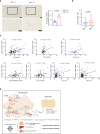
G-protein-signaling modulator 1 (GPSM1) exhibits strong genetic association with Type 2 diabetes (T2D) and Body Mass Index in population studies. However, how GPSM1 carries out such control and in which types of cells are poorly understood. Here, we demonstrate that myeloid GPSM1 promotes metabolic inflammation to accelerate T2D and obesity development. Mice with myeloid-specific GPSM1 ablation are protected against high fat diet-induced insulin resistance, glucose dysregulation, and liver steatosis via repression of adipose tissue pro-inflammatory states. Mechanistically, GPSM1 deficiency mainly promotes TNFAIP3 transcription via the Gαi3/cAMP/PKA/CREB axis, thus inhibiting TLR4-induced NF-κB signaling in macrophages. In addition, we identify a small-molecule compound, AN-465/42243987, which suppresses the pro-inflammatory phenotype by inhibiting GPSM1 function, which could make it a candidate for metabolic therapy. Furthermore, GPSM1 expression is upregulated in visceral fat of individuals with obesity and is correlated with clinical metabolic traits. Overall, our findings identify macrophage GPSM1 as a link between metabolic inflammation and systemic homeostasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

