Analysis of serum circulating MicroRNAs level in Malaysian patients with gestational diabetes mellitus
- PMID: 36434110
- PMCID: PMC9700700
- DOI: 10.1038/s41598-022-23816-3
Analysis of serum circulating MicroRNAs level in Malaysian patients with gestational diabetes mellitus
Erratum in
-
Author Correction: Analysis of serum circulating MicroRNAs level in Malaysian patients with gestational diabetes mellitus.Sci Rep. 2023 Mar 29;13(1):5121. doi: 10.1038/s41598-023-32260-w. Sci Rep. 2023. PMID: 36991146 Free PMC article. No abstract available.
Abstract
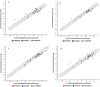
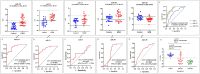
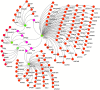
Gestational diabetes mellitus (GDM) is a severe global issue that requires immediate attention. MicroRNA expression abnormalities are possibly disease-specific and may contribute to GDM pathological processes. To date, there is limited data on miRNA profiling in GDM, especially that involves a longitudinal study. Here, we performed miRNA expression profiling in the entire duration of pregnancy (during pregnancy until parturition and postpartum) using a miRNA- polymerase chain reaction array (miRNA-PCRArray) and in-silico analysis to identify unique miRNAs expression and their anticipated target genes in Malay maternal serum. MiRNA expression levels and their unique potential as biomarkers were explored in this work. In GDM patients, the expression levels of hsa-miR-193a, hsa-miR-21, hsa-miR-23a, and hsa-miR-361 were significantly increased, but miR-130a was significantly downregulated. The area under the curve (AUC) and receiver operating characteristic (ROC) curve study demonstrated that hsa-miR-193a (AUC = 0.89060 ± 04,470, P = 0.0001), hsa-miR-21 (AUC = 0.89500 ± 04,411, P = 0.0001), and miR-130a (AUC = 0.6939 ± 0.05845, P = 0.0025) had potential biomarker features in GDM. In-silico analysis also revealed that KLF (Kruppel-Like family of transcription factor), ZNF25 (Zinc finger protein 25), AFF4 (ALF transcription elongation factor 4), C1orf143 (long intergenic non-protein coding RNA 2869), SRSF2 (serine and arginine rich splicing factor 2), and ZNF655 (Zinc finger protein 655) were prominent genes targeted by the common nodes of miR23a, miR130, miR193a, miR21, and miR361.Our findings suggest that circulating microRNAs in the first trimester has the potential for GDM screening in the Malay population.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients.BMC Res Notes. 2013 Nov 26;6:491. doi: 10.1186/1756-0500-6-491. BMC Res Notes. 2013. PMID: 24279768 Free PMC article.
-
Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes.Clin Epigenetics. 2017 Dec 12;9:127. doi: 10.1186/s13148-017-0428-1. eCollection 2017. Clin Epigenetics. 2017. PMID: 29255496 Free PMC article.
-
Circulating microRNAs associated with gestational diabetes mellitus: useful biomarkers?J Endocrinol. 2022 Dec 13;256(1):e220170. doi: 10.1530/JOE-22-0170. Print 2023 Jan 1. J Endocrinol. 2022. PMID: 36346274 Review.
-
The Predictive Value of miR-16, -29a and -134 for Early Identification of Gestational Diabetes: A Nested Analysis of the DALI Cohort.Cells. 2021 Jan 15;10(1):170. doi: 10.3390/cells10010170. Cells. 2021. PMID: 33467738 Free PMC article.
-
A Systematic Review of MicroRNA Expression as Biomarker of Late-Onset Alzheimer's Disease.Mol Neurobiol. 2019 Dec;56(12):8376-8391. doi: 10.1007/s12035-019-01676-9. Epub 2019 Jun 25. Mol Neurobiol. 2019. PMID: 31240600
Cited by
-
The role of ncRNA regulatory mechanisms in diseases-case on gestational diabetes.Brief Bioinform. 2023 Nov 22;25(1):bbad489. doi: 10.1093/bib/bbad489. Brief Bioinform. 2023. PMID: 38189542 Free PMC article. Review.
-
Circulating MicroRNAs Associated with Changes in the Placenta and Their Possible Role in the Fetus During Gestational Diabetes Mellitus: A Review.Metabolites. 2025 Jun 3;15(6):367. doi: 10.3390/metabo15060367. Metabolites. 2025. PMID: 40559391 Free PMC article. Review.
-
Global DNA methylation and miR-126-3p expression in Mexican women with gestational diabetes mellitus: a pilot study.Mol Biol Rep. 2023 Dec 12;51(1):5. doi: 10.1007/s11033-023-09005-z. Mol Biol Rep. 2023. PMID: 38085382
-
Upregulation of the Antioxidant Response-Related microRNAs miR-146a-5p and miR-21-5p in Gestational Diabetes: An Analysis of Matched Samples of Extracellular Vesicles and PBMCs.Int J Mol Sci. 2025 Jul 18;26(14):6902. doi: 10.3390/ijms26146902. Int J Mol Sci. 2025. PMID: 40725161 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

