Molecular basis of diseases induced by the mitochondrial DNA mutation m.9032T>C
- PMID: 36434790
- PMCID: PMC10077503
- DOI: 10.1093/hmg/ddac292
Molecular basis of diseases induced by the mitochondrial DNA mutation m.9032T>C
Abstract
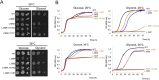
The mitochondrial DNA mutation m.9032T>C was previously identified in patients presenting with NARP (Neuropathy Ataxia Retinitis Pigmentosa). Their clinical features had a maternal transmission and patient's cells showed a reduced oxidative phosphorylation capacity, elevated reactive oxygen species (ROS) production and hyperpolarization of the mitochondrial inner membrane, providing evidence that m.9032T>C is truly pathogenic. This mutation leads to replacement of a highly conserved leucine residue with proline at position 169 of ATP synthase subunit a (L169P). This protein and a ring of identical c-subunits (c-ring) move protons through the mitochondrial inner membrane coupled to ATP synthesis. We herein investigated the consequences of m.9032T>C on ATP synthase in a strain of Saccharomyces cerevisiae with an equivalent mutation (L186P). The mutant enzyme assembled correctly but was mostly inactive as evidenced by a > 95% drop in the rate of mitochondrial ATP synthesis and absence of significant ATP-driven proton pumping across the mitochondrial membrane. Intragenic suppressors selected from L186P yeast restoring ATP synthase function to varying degrees (30-70%) were identified at the original mutation site (L186S) or in another position of the subunit a (H114Q, I118T). In light of atomic structures of yeast ATP synthase recently described, we conclude from these results that m.9032T>C disrupts proton conduction between the external side of the membrane and the c-ring, and that H114Q and I118T enable protons to access the c-ring through a modified pathway.
© The Author(s) 2022. Published by Oxford University Press.
Figures




References
-
- Saraste, M. (1999) Oxidative phosphorylation at the fin de siècle. Science, 283, 1488–1493. - PubMed
-
- DiMauro, S. and Schon, E.A. (2003) Mitochondrial respiratory-chain diseases. N. Engl. J. Med., 348, 2656–2668. - PubMed
-
- Vafai, S.B. and Mootha, V.K. (2012) Mitochondrial disorders as windows into an ancient organelle. Nature, 491, 374–383. - PubMed
-
- Zeviani, M. and Carelli, V. (2007) Mitochondrial disorders. Curr. Opin. Neurol., 20, 564–571. - PubMed
-
- Wallace, D.C. (2010) Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen., 51, 440–450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

