Letrozole treatment alters hippocampal gene expression in common marmosets (Callithrix jacchus)
- PMID: 36434852
- PMCID: PMC9839488
- DOI: 10.1016/j.yhbeh.2022.105281
Letrozole treatment alters hippocampal gene expression in common marmosets (Callithrix jacchus)
Abstract
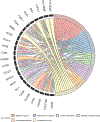
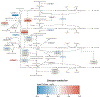
Aromatase inhibitors (AIs) are a class of drugs commonly given to patients with estrogen receptor (ER)-dependent breast cancers to reduce estrogenic stimulation. However, AIs like Letrozole are associated with negative side effects such as cognitive deficits, sleep disturbances and hot flashes. We have previously shown that these negative effects can be recapitulated in common marmosets (Callithrix jacchus) treated with Letrozole (20 μg daily) for 4 weeks and that marmosets treated with Letrozole show increased levels of estradiol in the hippocampus (Gervais et al., 2019). In order to better understand the mechanisms through which AIs affect cognitive function and increase steroid levels in the hippocampus, we used bulk, paired-end RNA-sequencing to examine differentially expressed genes among Letrozole-treated (LET; n = 8) and vehicle-treated (VEH; n = 8) male and female animals. Gene ontology results show significant reduction across hundreds of categories, some of the most significant being inflammatory response, stress response, MHC Class II protein complex binding, T-cell activation, carbohydrate binding and signaling receptor binding in LET animals. GSEA results indicate that LET females, but not LET males, show enrichment for hormonal gene sets. Based on the transcriptional changes observed, we conclude that AIs may differentially affect the sexes in part due to processes mediated by the CYP-450 superfamily. Ongoing studies will further investigate the longitudinal effects of AIs on behavior and whether AIs increase the risk of stress-induced neurodegeneration.
Keywords: Aromatase inhibitor; Estradiol; Hippocampus; Letrozole; RNA-sequencing.
Copyright © 2022 Elsevier Inc. All rights reserved.
Figures




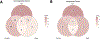



Similar articles
-
Adverse Effects of Aromatase Inhibition on the Brain and Behavior in a Nonhuman Primate.J Neurosci. 2019 Jan 30;39(5):918-928. doi: 10.1523/JNEUROSCI.0353-18.2018. Epub 2018 Dec 26. J Neurosci. 2019. PMID: 30587540 Free PMC article.
-
Extraovarian gonadotropin negative feedback revealed by aromatase inhibition in female marmoset monkeys.Am J Physiol Endocrinol Metab. 2017 Nov 1;313(5):E507-E514. doi: 10.1152/ajpendo.00058.2017. Epub 2017 Jul 5. Am J Physiol Endocrinol Metab. 2017. PMID: 28679622 Free PMC article.
-
Sex differences in behavioral and neurochemical effects of gonadectomy and aromatase inhibition in rats.Psychoneuroendocrinology. 2018 Jan;87:93-107. doi: 10.1016/j.psyneuen.2017.10.007. Epub 2017 Oct 10. Psychoneuroendocrinology. 2018. PMID: 29054014
-
Aromatase inhibitors and breast cancer.Ann N Y Acad Sci. 2009 Feb;1155:162-73. doi: 10.1111/j.1749-6632.2008.03689.x. Ann N Y Acad Sci. 2009. PMID: 19250202 Review.
-
The pharmacology of letrozole.J Steroid Biochem Mol Biol. 2003 Oct;87(1):35-45. doi: 10.1016/s0960-0760(03)00384-4. J Steroid Biochem Mol Biol. 2003. PMID: 14630089 Review.
Cited by
-
Letrozole delays acquisition of water maze task in female BALB/c mice: Possible involvement of anxiety.Horm Behav. 2024 Jun;162:105524. doi: 10.1016/j.yhbeh.2024.105524. Epub 2024 Mar 21. Horm Behav. 2024. PMID: 38513526 Free PMC article.
References
-
- Arora P, Gudelsky G, & Desai PB (2021). Gender-based differences in brain and plasma pharmacokinetics of letrozole in sprague-dawley rats: Application of physiologically-based pharmacokinetic modeling to gain quantitative insights. PLOS ONE, 16(4), e0248579. 10.1371/journal.pone.0248579 - DOI - PMC - PubMed
-
- Bian C, Zhao Y, Guo Q, Xiong Y, Cai W, & Zhang J (2014). Aromatase inhibitor letrozole downregulates steroid receptor coactivator-1 in specific brain regions that primarily related to memory, neuroendocrine and integration. The Journal of Steroid Biochemistry and Molecular Biology, 141, 37–43. 10.1016/j.jsbmb.2013.12.020 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

