Sample size requirement for achieving multisite harmonization using structural brain MRI features
- PMID: 36435343
- PMCID: PMC7615107
- DOI: 10.1016/j.neuroimage.2022.119768
Sample size requirement for achieving multisite harmonization using structural brain MRI features
Abstract
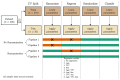
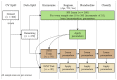
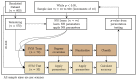
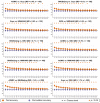
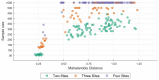
When data is pooled across multiple sites, the extracted features are confounded by site effects. Harmonization methods attempt to correct these site effects while preserving the biological variability within the features. However, little is known about the sample size requirement for effectively learning the harmonization parameters and their relationship with the increasing number of sites. In this study, we performed experiments to find the minimum sample size required to achieve multisite harmonization (using neuroHarmonize) using volumetric and surface features by leveraging the concept of learning curves. Our first two experiments show that site-effects are effectively removed in a univariate and multivariate manner; however, it is essential to regress the effect of covariates from the harmonized data additionally. Our following two experiments with actual and simulated data showed that the minimum sample size required for achieving harmonization grows with the increasing average Mahalanobis distances between the sites and their reference distribution. We conclude by positing a general framework to understand the site effects using the Mahalanobis distance. Further, we provide insights on the various factors in a cross-validation design to achieve optimal inter-site harmonization.
Keywords: Cross-validation; Harmonization; Mahalanobis distance; Multisite; Neuroimaging; Sample size.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest None
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

