Structural evolution of Delta lineage of SARS-CoV-2
- PMID: 36435470
- PMCID: PMC9683856
- DOI: 10.1016/j.ijbiomac.2022.11.227
Structural evolution of Delta lineage of SARS-CoV-2
Abstract
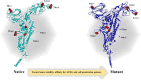
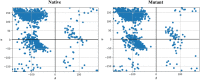
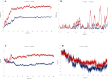
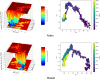
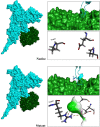
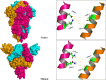
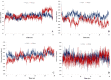
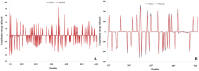
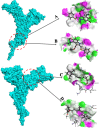
One of the main obstacles in prevention and treatment of COVID-19 is the rapid evolution of the SARS-CoV-2 Spike protein. Given that Spike is the main target of common treatments of COVID-19, mutations occurring at this virulent factor can affect the effectiveness of treatments. The B.1.617.2 lineage of SARS-CoV-2, being characterized by many Spike mutations inside and outside of its receptor-binding domain (RBD), shows high infectivity and relative resistance to existing cures. Here, utilizing a wide range of computational biology approaches, such as immunoinformatics, molecular dynamics (MD), analysis of intrinsically disordered regions (IDRs), protein-protein interaction analyses, residue scanning, and free energy calculations, we examine the structural and biological attributes of the B.1.617.2 Spike protein. Furthermore, the antibody design protocol of Rosetta was implemented for evaluation the stability and affinity improvement of the Bamlanivimab (LY-CoV55) antibody, which is not capable of interactions with the B.1.617.2 Spike. We observed that the detected mutations in the Spike of the B1.617.2 variant of concern can cause extensive structural changes compatible with the described variation in immunogenicity, secondary and tertiary structure, oligomerization potency, Furin cleavability, and drug targetability. Compared to the Spike of Wuhan lineage, the B.1.617.2 Spike is more stable and binds to the Angiotensin-converting enzyme 2 (ACE2) with higher affinity.
Keywords: B.1.617.2; COVID-19; Computational biology; Mutation; SARS-CoV-2; Spike.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest Mohammad Mahmoudi Gomari reports financial support was provided by Iran University of Medical Sciences.
Figures






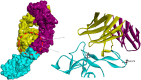
Similar articles
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
Computational modeling of the effect of five mutations on the structure of the ACE2 receptor and their correlation with infectivity and virulence of some emerged variants of SARS-CoV-2 suggests mechanisms of binding affinity dysregulation.Chem Biol Interact. 2022 Dec 1;368:110244. doi: 10.1016/j.cbi.2022.110244. Epub 2022 Nov 3. Chem Biol Interact. 2022. PMID: 36336003 Free PMC article.
-
Assessment of mutations on RBD in the Spike protein of SARS-CoV-2 Alpha, Delta and Omicron variants.Sci Rep. 2022 May 20;12(1):8540. doi: 10.1038/s41598-022-12479-9. Sci Rep. 2022. PMID: 35595778 Free PMC article.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
-
Mutations in the SARS-CoV-2 spike receptor binding domain and their delicate balance between ACE2 affinity and antibody evasion.Protein Cell. 2024 May 28;15(6):403-418. doi: 10.1093/procel/pwae007. Protein Cell. 2024. PMID: 38442025 Free PMC article. Review.
Cited by
-
Unleashing the potential of traditional Chinese medicine: a computational approach to discovering drug targets utilizing the CSLN and molecular dynamics.Mol Divers. 2025 May 3. doi: 10.1007/s11030-025-11177-8. Online ahead of print. Mol Divers. 2025. PMID: 40317435
-
Mutational dynamics of SARS-CoV-2: Impact on future COVID-19 vaccine strategies.Heliyon. 2024 Apr 25;10(9):e30208. doi: 10.1016/j.heliyon.2024.e30208. eCollection 2024 May 15. Heliyon. 2024. PMID: 38707429 Free PMC article. Review.
-
QSAR, ADMET, molecular docking, and dynamics studies of 1,2,4-triazine-3(2H)-one derivatives as tubulin inhibitors for breast cancer therapy.Sci Rep. 2024 Jul 16;14(1):16418. doi: 10.1038/s41598-024-66877-2. Sci Rep. 2024. PMID: 39013949 Free PMC article.
-
Potent VEGFR-2 inhibitors for resistant breast cancer: a comprehensive 3D-QSAR, ADMET, molecular docking and MMPBSA calculation on triazolopyrazine derivatives.Front Mol Biosci. 2023 Nov 22;10:1288652. doi: 10.3389/fmolb.2023.1288652. eCollection 2023. Front Mol Biosci. 2023. PMID: 38074087 Free PMC article.
-
Design, Synthesis, and Comparison of PLA-PEG-PLA and PEG-PLA-PEG Copolymers for Curcumin Delivery to Cancer Cells.Polymers (Basel). 2023 Jul 23;15(14):3133. doi: 10.3390/polym15143133. Polymers (Basel). 2023. PMID: 37514522 Free PMC article.
References
-
- COVID-19 Dashboard Coronavirus resource center. 2021. https://coronavirus.jhu.edu/map.html Available from:
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

