Structural characterization of protective non-neutralizing antibodies targeting Crimean-Congo hemorrhagic fever virus
- PMID: 36435827
- PMCID: PMC9701186
- DOI: 10.1038/s41467-022-34923-0
Structural characterization of protective non-neutralizing antibodies targeting Crimean-Congo hemorrhagic fever virus
Abstract
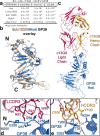
Crimean-Congo Hemorrhagic Fever Virus (CCHFV) causes a life-threatening disease with up to a 40% mortality rate. With no approved medical countermeasures, CCHFV is considered a public health priority agent. The non-neutralizing mouse monoclonal antibody (mAb) 13G8 targets CCHFV glycoprotein GP38 and protects mice from lethal CCHFV challenge when administered prophylactically or therapeutically. Here, we reveal the structures of GP38 bound with a human chimeric 13G8 mAb and a newly isolated CC5-17 mAb from a human survivor. These mAbs bind overlapping epitopes with a shifted angle. The broad-spectrum potential of c13G8 and CC5-17 and the practicality of using them against Aigai virus, a closely related nairovirus were examined. Binding studies demonstrate that the presence of non-conserved amino acids in Aigai virus corresponding region prevent CCHFV mAbs from binding Aigai virus GP38. This information, coupled with in vivo efficacy, paves the way for future mAb therapeutics effective against a wide swath of CCHFV strains.
© 2022. The Author(s).
Conflict of interest statement
A.R.G. and J.W.G. are inventors on a patent application related to the use of mAb 13G8 as a therapeutic, PCT/US2020/012621. The remaining authors declare no competing interests.
Figures


Similar articles
-
Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38.J Virol. 2020 Mar 31;94(8):e02005-19. doi: 10.1128/JVI.02005-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996434 Free PMC article.
-
GP38-targeting monoclonal antibodies protect adult mice against lethal Crimean-Congo hemorrhagic fever virus infection.Sci Adv. 2019 Jul 10;5(7):eaaw9535. doi: 10.1126/sciadv.aaw9535. eCollection 2019 Jul. Sci Adv. 2019. PMID: 31309159 Free PMC article.
-
Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.Cell Rep. 2024 Jul 23;43(7):114502. doi: 10.1016/j.celrep.2024.114502. Epub 2024 Jul 13. Cell Rep. 2024. PMID: 39002130 Free PMC article.
-
Crimean-Congo Hemorrhagic Fever.Lab Med. 2015 Summer;46(3):180-9. doi: 10.1309/LMN1P2FRZ7BKZSCO. Lab Med. 2015. PMID: 26199256 Review.
-
Crimean-Congo haemorrhagic fever virus: Past, present and future insights for animal modelling and medical countermeasures.Zoonoses Public Health. 2018 Aug;65(5):465-480. doi: 10.1111/zph.12469. Epub 2018 Apr 20. Zoonoses Public Health. 2018. PMID: 29676526 Free PMC article. Review.
Cited by
-
Neutralizing monoclonal antibodies against the Gc fusion loop region of Crimean-Congo hemorrhagic fever virus.PLoS Pathog. 2024 Feb 1;20(2):e1011948. doi: 10.1371/journal.ppat.1011948. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38300972 Free PMC article.
-
Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers.Commun Biol. 2025 May 29;8(1):825. doi: 10.1038/s42003-025-08239-w. Commun Biol. 2025. PMID: 40442315 Free PMC article.
-
Crimean Congo hemorrhagic fever virus nucleoprotein and GP38 subunit vaccine combination prevents morbidity in mice.NPJ Vaccines. 2024 Aug 14;9(1):148. doi: 10.1038/s41541-024-00931-y. NPJ Vaccines. 2024. PMID: 39143104 Free PMC article.
-
Nucleocapsid protein-specific monoclonal antibodies protect mice against Crimean-Congo hemorrhagic fever virus.Nat Commun. 2024 Feb 26;15(1):1722. doi: 10.1038/s41467-024-46110-4. Nat Commun. 2024. PMID: 38409240 Free PMC article.
-
The Adaptive Immune Response against Bunyavirales.Viruses. 2024 Mar 21;16(3):483. doi: 10.3390/v16030483. Viruses. 2024. PMID: 38543848 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

