Persistent inflammation and neuronal loss in the mouse brain induced by a modified form of attenuated herpes simplex virus type I
- PMID: 36436797
- PMCID: PMC10006190
- DOI: 10.1016/j.virs.2022.11.008
Persistent inflammation and neuronal loss in the mouse brain induced by a modified form of attenuated herpes simplex virus type I
Abstract
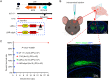
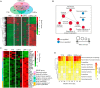
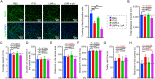
Herpes simplex virus-1 (HSV-1) is a widespread neurotropic virus that can reach the brain and cause a rare but acute herpes simplex encephalitis (HSE) with a high mortality rate. Most patients present with changes in neurological and behavioral status, and survivors suffer long-term neurological sequelae. To date, the pathogenesis leading to brain damage is still not well understood. HSV-1 induced encephalitis in the central nervous system (CNS) in animals are usually very diffuse and progressing rapidly, and mostly fatal, making the analysis difficult. Here, we established a mouse model of HSE via intracerebral inoculation of modified version of neural-attenuated strains of HSV-1 (deletion of ICP34.5 and inserting a strong promoter into the latency-associated transcript region), in which the LMR-αΔpA strain initiated moderate productive infection, leading to strong host immune and inflammatory response characterized by persistent microglia activation. This viral replication activity and prolonged inflammatory response activated signaling pathways in neuronal damage, amyloidosis, Alzheimer's disease, and neurodegeneration, eventually leading to neuronal loss and behavioral changes characterized by hypokinesia. Our study reveals detailed pathogenic processes and persistent inflammatory responses in the CNS and provides a controlled, mild and non-lethal HSE model for studying long-term neuronal injury and increased risk of neurodegenerative diseases due to HSV-1 infection.
Keywords: Herpes simplex encephalitis (HSE); Herpes simplex virus-1 (HSV-1); ICP34.5; Neuroinflammation; Neuronal loss.
Copyright © 2022 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Microglia Activate Early Antiviral Responses upon Herpes Simplex Virus 1 Entry into the Brain to Counteract Development of Encephalitis-Like Disease in Mice.J Virol. 2022 Mar 23;96(6):e0131121. doi: 10.1128/JVI.01311-21. Epub 2022 Jan 19. J Virol. 2022. PMID: 35045263 Free PMC article.
-
Ketogenic diet restrains herpes simplex encephalitis via gut microbes.Microbes Infect. 2023 Mar-Apr;25(3):105061. doi: 10.1016/j.micinf.2022.105061. Epub 2022 Oct 18. Microbes Infect. 2023. PMID: 36270600
-
Microglial mitophagy integrates the microbiota-gut-brain axis to restrain neuroinflammation during neurotropic herpesvirus infection.Autophagy. 2023 Feb;19(2):734-736. doi: 10.1080/15548627.2022.2102309. Epub 2022 Jul 23. Autophagy. 2023. PMID: 35849507 Free PMC article.
-
Herpes simplex virus infections of the central nervous system: encephalitis and meningitis, including Mollaret's.Herpes. 2004 Jun;11 Suppl 2:57A-64A. Herpes. 2004. PMID: 15319091 Review.
-
Innate immune response during herpes simplex virus encephalitis and development of immunomodulatory strategies.Rev Med Virol. 2015 Sep;25(5):300-19. doi: 10.1002/rmv.1848. Epub 2015 Jul 23. Rev Med Virol. 2015. PMID: 26205506 Review.
Cited by
-
Biological agents and the aging brain: glial inflammation and neurotoxic signaling.Front Aging. 2023 Aug 15;4:1244149. doi: 10.3389/fragi.2023.1244149. eCollection 2023. Front Aging. 2023. PMID: 37649972 Free PMC article. Review.
-
Microbial infection promotes amyloid pathology in a mouse model of Alzheimer's disease via modulating γ-secretase.Mol Psychiatry. 2024 May;29(5):1491-1500. doi: 10.1038/s41380-024-02428-5. Epub 2024 Jan 25. Mol Psychiatry. 2024. PMID: 38273109
References
-
- Asai M., Hattori C., Szabo B., Sasagawa N., Maruyama K., Tanuma S., Ishiura S. Putative function of adam9, adam10, and adam17 as app alpha-secretase. Biochem. Biophys. Res. Commun. 2003;301:231–235. - PubMed
-
- Barker K.R., Sarafino-Wani R., Khanom A., Griffiths P.D., Jacobs M.G., Webster D.P. Encephalitis in an immunocompetent man. J. Clin. Virol. 2014;59:1–3. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

