Novel candidate loci for morpho-agronomic and seed quality traits detected by targeted genotyping-by-sequencing in common bean
- PMID: 36438107
- PMCID: PMC9685177
- DOI: 10.3389/fpls.2022.1014282
Novel candidate loci for morpho-agronomic and seed quality traits detected by targeted genotyping-by-sequencing in common bean
Abstract
Phaseolus vulgaris L., known as common bean, is one of the most important grain legumes cultivated around the world for its immature pods and dry seeds, which are rich in protein and micronutrients. Common bean offers a cheap food and protein sources to ameliorate food shortage and malnutrition around the world. However, the genetic basis of most important traits in common bean remains unknown. This study aimed at identifying QTL and candidate gene models underlying twenty-six agronomically important traits in common bean. For this, we assembled and phenotyped a diversity panel of 200 P. vulgaris genotypes in the greenhouse, comprising determinate bushy, determinate climbing and indeterminate climbing beans. The panel included dry beans and snap beans from different breeding programmes, elite lines and landraces from around the world with a major focus on accessions of African, European and South American origin. The panel was genotyped using a cost-conscious targeted genotyping-by-sequencing (GBS) platform to take advantage of highly polymorphic SNPs detected in previous studies and in diverse germplasm. The detected single nucleotide polymorphisms (SNPs) were applied in marker-trait analysis and revealed sixty-two quantitative trait loci (QTL) significantly associated with sixteen traits. Gene model identification via a similarity-based approach implicated major candidate gene models underlying the QTL associated with ten traits including, flowering, yield, seed quality, pod and seed characteristics. Our study revealed six QTL for pod shattering including three new QTL potentially useful for breeding. However, the panel was evaluated in a single greenhouse environment and the findings should be corroborated by evaluations across different field environments. Some of the detected QTL and a number of candidate gene models only elucidate the understanding of the genetic nature of these traits and provide the basis for further studies. Finally, the study showed the possibility of using a limited number of SNPs in performing marker-trait association in common bean by applying a highly scalable targeted GBS approach. This targeted GBS approach is a cost-efficient strategy for assessment of the genetic basis of complex traits and can enable geneticists and breeders to identify novel loci and targets for marker-assisted breeding more efficiently.
Keywords: GWAS; Phaseolus vulgaris; marker-trait association; phenology; pod shattering; seed quality; single nucleotide polymorphisms; targeted genotyping-by-sequencing.
Copyright © 2022 Ugwuanyi, Udengwu, Snowdon and Obermeier.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Figures


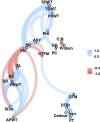

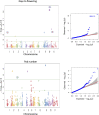

References
-
- Adesoye A. I., Ojobo O. A. (2012). Genetic diversity assessment of phaseolus vulgaris l. landraces in nigeria’s mid-altitude agroecological zone. int. J. Biodivers. Conserv. 4, 453–460. doi: 10.5897/IJBC11.216 - DOI
-
- Ahmad K. M. S. (2018). Genetic diversity of common bean (Phaseolus vulgaris) cultivars from different origins revealed by microsatellite markers. J. Adv. Biol. Biotechnol. 174, 1–9. doi: 10.9734/JABB/2018/40779 - DOI
-
- Ambachew D., Mekbib F., Asfaw A., Beebe S. E., Blair M. W. (2015). Trait associations in common bean genotypes grown under drought stress and field infestation by BSM bean fly. Crop J. 3, 305–316. doi: 10.1016/j.cj.2015.01.006 - DOI
-
- AOAC (1990). Official methods of analysis. 15th ed (Arlington, VA, USA: Association of Official Analytical Chemists; ).
-
- Assefa T., Mahama A. A., Brown A. V., Cannon E. K. S., Rubyogo J. C., Rao I. M., et al. . (2019). A review of breeding objectives, genomic resources and marker-assisted methods in common bean (Phaseolus vulgaris l.). Mol. Breed. 39, 20. doi: 10.1007/s11032-018-0920-0 - DOI
LinkOut - more resources
Full Text Sources

