A graph-based approach for simultaneous semantic and instance segmentation of plant 3D point clouds
- PMID: 36438118
- PMCID: PMC9691340
- DOI: 10.3389/fpls.2022.1012669
A graph-based approach for simultaneous semantic and instance segmentation of plant 3D point clouds
Abstract
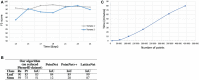
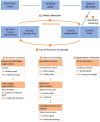
Accurate simultaneous semantic and instance segmentation of a plant 3D point cloud is critical for automatic plant phenotyping. Classically, each organ of the plant is detected based on the local geometry of the point cloud, but the consistency of the global structure of the plant is rarely assessed. We propose a two-level, graph-based approach for the automatic, fast and accurate segmentation of a plant into each of its organs with structural guarantees. We compute local geometric and spectral features on a neighbourhood graph of the points to distinguish between linear organs (main stem, branches, petioles) and two-dimensional ones (leaf blades) and even 3-dimensional ones (apices). Then a quotient graph connecting each detected macroscopic organ to its neighbors is used both to refine the labelling of the organs and to check the overall consistency of the segmentation. A refinement loop allows to correct segmentation defects. The method is assessed on both synthetic and real 3D point-cloud data sets of Chenopodium album (wild spinach) and Solanum lycopersicum (tomato plant).
Keywords: Fiedler vector; instance segmentation; phenotyping; quotient graph; semantic segmentation.
Copyright © 2022 Mirande, Godin, Tisserand, Charlaix, Besnard and Hétroy-Wheeler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- CloudCompare (2022). “GPL Software,” in Cloudcompare.
-
- Bengfort B., Bilbro R., Danielsen N., Gray L., McIntyre K., Roman P., et al. . Yellowbrick.(2018) doi: 10.5281/zenodo.1206264 - DOI
LinkOut - more resources
Full Text Sources

