The landscape of cancer-associated fibroblasts in colorectal cancer liver metastases
- PMID: 36438498
- PMCID: PMC9691344
- DOI: 10.7150/thno.72853
The landscape of cancer-associated fibroblasts in colorectal cancer liver metastases
Abstract
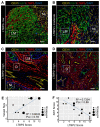
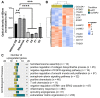
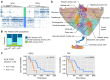
Rationale: Patients with colorectal cancer die mainly due to liver metastases (CRC-LM). Although the tumor microenvironment (TME) plays an important role in tumor development and therapeutic response, our understanding of the individual TME components, especially cancer-associated fibroblasts (CAFs), remains limited. Methods: We analyzed CRC-LM CAFs and cancer cells by single-cell transcriptomics and used bioinformatics for data analysis and integration with related available single-cell and bulk transcriptomic datasets. We validated key findings by RT-qPCR, western blotting, and immunofluorescence. Results: By single-cell transcriptomic analysis of 4,397 CAFs from six CRC-LM samples, we identified two main CAF populations, contractile CAFs and extracellular matrix (ECM)-remodeling/pro-angiogenic CAFs, and four subpopulations with distinct phenotypes. We found that ECM-remodeling/pro-angiogenic CAFs derive from portal resident fibroblasts. They associate with areas of strong desmoplastic reaction and Wnt signaling in low-proliferating tumor cells engulfed in a stiff extracellular matrix. By integrating public single-cell primary liver tumor data, we propose a model to explain how different liver malignancies recruit CAFs of different origins to this organ. Lastly, we found that LTBP2 plays an important role in modulating collagen biosynthesis, ECM organization, and adhesion pathways. We developed fully human antibodies against LTBP2 that depleted LTBP2+ CAFs in vitro. Conclusion: This study complements recent reports on CRC-LM CAF heterogeneity at the single-cell resolution. The number of sequenced CAFs was more than one order of magnitude larger compared to existing data. LTBP2 targeting by antibodies might create opportunities to deplete ECM-remodeling CAFs in CRC-LMs. This might be combined with other therapies, e.g., anti-angiogenic compounds as already done in CRC. Moreover, we showed that in intrahepatic cholangiocarcinoma, in which ECM-remodeling CAF proportion is similar to that of CRC-LM, several genes expressed by ECM-remodeling CAFs, such as LTBP2, were associated with survival.
Keywords: CAF; LTBP2.; liver metastasis; single-cell; tumor microenvironment.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80. - PubMed
-
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. - PubMed
-
- Emambux S, Tachon G, Junca A, Tougeron D. Results and challenges of immune checkpoint inhibitors in colorectal cancer. Expert Opin Biol Ther. 2018;18:561–73. - PubMed
-
- Ronca R, Van Ginderachter JA, Turtoi A. Paracrine interactions of cancer-associated fibroblasts, macrophages and endothelial cells: tumor allies and foes. Curr Opin Oncol. 2018;30(1):45–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

