3-deazaadenosine (3DA) alleviates senescence to promote cellular fitness and cell therapy efficiency in mice
- PMID: 36438588
- PMCID: PMC7613850
- DOI: 10.1038/s43587-022-00279-9
3-deazaadenosine (3DA) alleviates senescence to promote cellular fitness and cell therapy efficiency in mice
Abstract
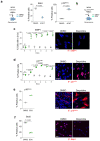
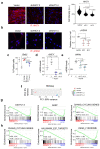
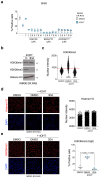
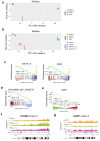
Cellular senescence is a stable type of cell cycle arrest triggered by different stresses. As such, senescence drives age-related diseases and curbs cellular replicative potential. Here, we show that 3-deazaadenosine (3DA), an S-adenosyl homocysteinase (AHCY) inhibitor, alleviates replicative and oncogene-induced senescence. 3DA-treated senescent cells showed reduced global Histone H3 Lysine 36 trimethylation (H3K36me3), an epigenetic modification that marks the bodies of actively transcribed genes. By integrating transcriptome and epigenome data, we demonstrate that 3DA treatment affects key factors of the senescence transcriptional program. Remarkably, 3DA treatment alleviated senescence and increased the proliferative and regenerative potential of muscle stem cells from very old mice in vitro and in vivo. Moreover, ex vivo 3DA treatment was sufficient to enhance the engraftment of human umbilical cord blood (UCB) cells in immunocompromised mice. Together, our results identify 3DA as a promising drug enhancing the efficiency of cellular therapies by restraining senescence.
Keywords: 3DA; AHCY; cord blood cells; muscle stem cells; senescence.
Conflict of interest statement
Competing Interests J.G. has acted as a consultant for Unity Biotechnology, Geras Bio, Myricx Pharma and Merck KGaA. Pfizer and Unity Biotechnology have funded research in J.G.’s lab (unrelated to the work presented here). J.G. owns equity in Geras Bio. J.G. and A.G. are named inventors in an MRC patent and J.G. is a named inventor in another Imperial College patents, both related to senolytic therapies (the patents are no related to the work presented here). T.W. is scientific co-founder of and holds stakes in Cargene Therapeutics, which develops nucleic-acid therapeutics for liver diseases (unrelated to the work presented here). The remaining authors declare no competing interests.
Figures

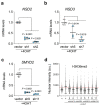


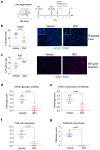



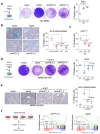
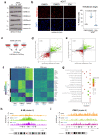


References
-
- Gorgoulis V, et al. Cellular Senescence: Defining a Path Forward. Cell. 2019;179:813–827. - PubMed
-
- Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. - PubMed
-
- Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

