Inventa: A computational tool to discover structural novelty in natural extracts libraries
- PMID: 36438653
- PMCID: PMC9692083
- DOI: 10.3389/fmolb.2022.1028334
Inventa: A computational tool to discover structural novelty in natural extracts libraries
Abstract
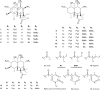
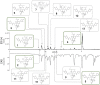
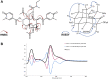
Collections of natural extracts hold potential for the discovery of novel natural products with original modes of action. The prioritization of extracts from collections remains challenging due to the lack of a workflow that combines multiple-source information to facilitate the data interpretation. Results from different analytical techniques and literature reports need to be organized, processed, and interpreted to enable optimal decision-making for extracts prioritization. Here, we introduce Inventa, a computational tool that highlights the structural novelty potential within extracts, considering untargeted mass spectrometry data, spectral annotation, and literature reports. Based on this information, Inventa calculates multiple scores that inform their structural potential. Thus, Inventa has the potential to accelerate new natural products discovery. Inventa was applied to a set of plants from the Celastraceae family as a proof of concept. The Pristimera indica (Willd.) A.C.Sm roots extract was highlighted as a promising source of potentially novel compounds. Its phytochemical investigation resulted in the isolation and de novo characterization of thirteen new dihydro-β-agarofuran sesquiterpenes, five of them presenting a new 9-oxodihydro-β-agarofuran base scaffold.
Keywords: bioinformatic tools; computational metabolomics; mass spectrometry; natural products; prioritization; structural novelty discovery.
Copyright © 2022 Quiros-Guerrero, Nothias, Gaudry, Marcourt, Allard, Rutz, David, Queiroz and Wolfender.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Alvarenga N., Ferro E. A. (2006). “Bioactive triterpenes and related compounds from celastraceae,” in Studies in natural products chemistry. Editor Atta-ur-Rahman (Elsevier; ), 239–307. 10.1016/S1572-5995(06)80029-3 - DOI
-
- Brejnrod A., Ernst M., Dworzynski P., Rasmussen L. B., Dorrestein P. C., van der Hooft J. J. J., et al. (2019). Implementations of the chemical structural and compositional similarity metric in R and Python. bioRxiv 1, 546150. 10.1101/546150 - DOI
LinkOut - more resources
Full Text Sources

