The mitochondrial Cu+ transporter PiC2 (SLC25A3) is a target of MTF1 and contributes to the development of skeletal muscle in vitro
- PMID: 36438658
- PMCID: PMC9682256
- DOI: 10.3389/fmolb.2022.1037941
The mitochondrial Cu+ transporter PiC2 (SLC25A3) is a target of MTF1 and contributes to the development of skeletal muscle in vitro
Abstract
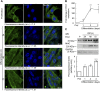
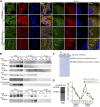
The loading of copper (Cu) into cytochrome c oxidase (COX) in mitochondria is essential for energy production in cells. Extensive studies have been performed to characterize mitochondrial cuproenzymes that contribute to the metallation of COX, such as Sco1, Sco2, and Cox17. However, limited information is available on the upstream mechanism of Cu transport and delivery to mitochondria, especially through Cu-impermeable membranes, in mammalian cells. The mitochondrial phosphate transporter SLC25A3, also known as PiC2, binds Cu+ and transports the ion through these membranes in eukaryotic cells, ultimately aiding in the metallation of COX. We used the well-established differentiation model of primary myoblasts derived from mouse satellite cells, wherein Cu availability is necessary for growth and maturation, and showed that PiC2 is a target of MTF1, and its expression is both induced during myogenesis and favored by Cu supplementation. PiC2 deletion using CRISPR/Cas9 showed that the transporter is required for proliferation and differentiation of primary myoblasts, as both processes are delayed upon PiC2 knock-out. The effects of PiC2 deletion were rescued by the addition of Cu to the growth medium, implying the deleterious effects of PiC2 knockout in myoblasts may be in part due to a failure to deliver sufficient Cu to the mitochondria, which can be compensated by other mitochondrial cuproproteins. Co-localization and co-immunoprecipitation of PiC2 and COX also suggest that PiC2 may participate upstream in the copper delivery chain into COX, as verified by in vitro Cu+-transfer experiments. These data indicate an important role for PiC2 in both the delivery of Cu to the mitochondria and COX, favoring the differentiation of primary myoblasts.
Keywords: MTF1; PiC2; SLC25A3; copper transport; cytochrome c oxidase; mitochondria.
Copyright © 2022 McCann, Quinteros, Adelugba, Morgada, Castelblanco, Davis, Lanzirotti, Hainer, Vila, Navea and Padilla-Benavides.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Andrews G. K., Lee D. K., Ravindra R., Lichtlen P., Sirito M., Sawadogo M., et al. (2001). The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J. 20, 1114–1122. 10.1093/emboj/20.5.1114 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

