Type I interferon receptor (IFNAR2) deficiency reveals Zika virus cytopathicity in human macrophages and microglia
- PMID: 36439115
- PMCID: PMC9691778
- DOI: 10.3389/fimmu.2022.1035532
Type I interferon receptor (IFNAR2) deficiency reveals Zika virus cytopathicity in human macrophages and microglia
Abstract
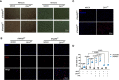
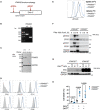
Macrophages are key target cells of Zika virus (ZIKV) infection, implicated as a viral reservoir seeding sanctuary sites such as the central nervous system and testes. This rests on the apparent ability of macrophages to sustain ZIKV replication without experiencing cytopathic effects. ZIKV infection of macrophages triggers an innate immune response involving type I interferons (IFN-I), key antiviral cytokines that play a complex role in ZIKV pathogenesis in animal models. To investigate the functional role of the IFN-I response we generated human induced pluripotent stem cell (iPSC)-derived macrophages from a patient with complete deficiency of IFNAR2, the high affinity IFN-I receptor subunit. Accompanying the profound defect of IFN-I signalling in IFNAR2 deficient iPS-macrophages we observed significantly enhanced ZIKV replication and cell death, revealing the inherent cytopathicity of ZIKV towards macrophages. These observations were recapitulated by genetic and pharmacological ablation of IFN-I signalling in control iPS-macrophages and extended to a model of iPS-microglia. Thus, the capacity of macrophages to support noncytolytic ZIKV replication depends on an equilibrium set by IFN-I, suggesting that innate antiviral responses might counterintuitively promote ZIKV persistence via the maintenance of tissue viral reservoirs relevant to pathogenesis.
Keywords: IFNAR2 deficiency; Zika virus; antiviral state; cell death; inborn errors of immunity; interferon-stimulated genes; macrophages; type I interferons.
Copyright © 2022 Hanrath, Hatton, Gothe, Browne, Vowles, Leary, Cockell, Cowley, James, Hambleton and Duncan.
Conflict of interest statement
SH declares honoraria from CSL Behring and Takeda for teaching and consultancy. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

