Circular RNAs as emerging regulators in COVID-19 pathogenesis and progression
- PMID: 36439162
- PMCID: PMC9681929
- DOI: 10.3389/fimmu.2022.980231
Circular RNAs as emerging regulators in COVID-19 pathogenesis and progression
Abstract
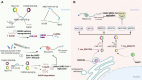
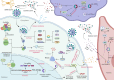
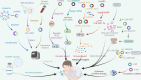
Coronavirus disease 2019 (COVID-19), an infectious acute respiratory disease caused by a newly emerging RNA virus, is a still-growing pandemic that has caused more than 6 million deaths globally and has seriously threatened the lives and health of people across the world. Currently, several drugs have been used in the clinical treatment of COVID-19, such as small molecules, neutralizing antibodies, and monoclonal antibodies. In addition, several vaccines have been used to prevent the spread of the pandemic, such as adenovirus vector vaccines, inactivated vaccines, recombinant subunit vaccines, and nucleic acid vaccines. However, the efficacy of vaccines and the onset of adverse reactions vary among individuals. Accumulating evidence has demonstrated that circular RNAs (circRNAs) are crucial regulators of viral infections and antiviral immune responses and are heavily involved in COVID-19 pathologies. During novel coronavirus infection, circRNAs not only directly affect the transcription process and interfere with viral replication but also indirectly regulate biological processes, including virus-host receptor binding and the immune response. Consequently, understanding the expression and function of circRNAs during severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection will provide novel insights into the development of circRNA-based methods. In this review, we summarize recent progress on the roles and underlying mechanisms of circRNAs that regulate the inflammatory response, viral replication, immune evasion, and cytokines induced by SARS-CoV-2 infection, and thus highlighting the diagnostic and therapeutic challenges in the treatment of COVID-19 and future research directions.
Keywords: COVID-19; biological regulator; circRNAs; inflammatory response; vaccine.
Copyright © 2022 Gao, Fang, Liang, Deng, Chen, Zeng and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Long Noncoding RNAs as Emerging Regulators of COVID-19.Front Immunol. 2021 Aug 2;12:700184. doi: 10.3389/fimmu.2021.700184. eCollection 2021. Front Immunol. 2021. PMID: 34408749 Free PMC article. Review.
-
Circulating microRNAs as emerging regulators of COVID-19.Theranostics. 2023 Jan 1;13(1):125-147. doi: 10.7150/thno.78164. eCollection 2023. Theranostics. 2023. PMID: 36593971 Free PMC article. Review.
-
Differential host circRNA expression profiles in human lung epithelial cells infected with SARS-CoV-2.Infect Genet Evol. 2021 Sep;93:104923. doi: 10.1016/j.meegid.2021.104923. Epub 2021 May 15. Infect Genet Evol. 2021. PMID: 34004360 Free PMC article.
-
hnRNP C modulates MERS-CoV and SARS-CoV-2 replication by governing the expression of a subset of circRNAs and cognitive mRNAs.Emerg Microbes Infect. 2022 Dec;11(1):519-531. doi: 10.1080/22221751.2022.2032372. Emerg Microbes Infect. 2022. PMID: 35060842 Free PMC article.
-
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants.Elife. 2022 Aug 25;11:e78633. doi: 10.7554/eLife.78633. Elife. 2022. PMID: 36004719 Free PMC article.
Cited by
-
Altered circular RNA expressions in extracellular vesicles from bronchoalveolar lavage fluids in mice after bacterial infections.Front Immunol. 2024 Apr 4;15:1354676. doi: 10.3389/fimmu.2024.1354676. eCollection 2024. Front Immunol. 2024. PMID: 38638425 Free PMC article.
-
Simultaneous effect of different chromatographic conditions on the chromatographic retention of pentapeptide derivatives (HGRFG and NPNPT).Front Chem. 2023 Apr 18;11:1171824. doi: 10.3389/fchem.2023.1171824. eCollection 2023. Front Chem. 2023. PMID: 37143822 Free PMC article.
-
Advances research in porcine enteric coronavirus therapies and antiviral drugs.Vet Q. 2024 Dec;44(1):1-49. doi: 10.1080/01652176.2024.2421299. Epub 2024 Nov 1. Vet Q. 2024. PMID: 39484691 Free PMC article. Review.
-
Non-coding RNAs expression in SARS-CoV-2 infection: pathogenesis, clinical significance, and therapeutic targets.Signal Transduct Target Ther. 2023 Dec 6;8(1):441. doi: 10.1038/s41392-023-01669-0. Signal Transduct Target Ther. 2023. PMID: 38057315 Free PMC article. Review.
-
Stimulation of PSTPIP1 to trigger proinflammatory responses in asymptomatic SARS-CoV-2 infections.Heliyon. 2024 Feb 28;10(5):e26886. doi: 10.1016/j.heliyon.2024.e26886. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38463809 Free PMC article.
References
-
- Aleem A, Akbar Samad AB, Slenker AK. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19). StatPearls. Treasure Island (FL: StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC; (2022). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

